Tracking Cancer Evolution through the Disease Course
- PMID: 33811124
- PMCID: PMC7611362
- DOI: 10.1158/2159-8290.CD-20-1559
Tracking Cancer Evolution through the Disease Course
Abstract
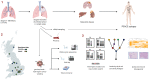
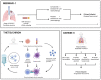
During cancer evolution, constituent tumor cells compete under dynamic selection pressures. Phenotypic variation can be observed as intratumor heterogeneity, which is propagated by genome instability leading to mutations, somatic copy-number alterations, and epigenomic changes. TRACERx was set up in 2014 to observe the relationship between intratumor heterogeneity and patient outcome. By integrating multiregion sequencing of primary tumors with longitudinal sampling of a prospectively recruited patient cohort, cancer evolution can be tracked from early- to late-stage disease and through therapy. Here we review some of the key features of the studies and look to the future of the field. SIGNIFICANCE: Cancers evolve and adapt to environmental challenges such as immune surveillance and treatment pressures. The TRACERx studies track cancer evolution in a clinical setting, through primary disease to recurrence. Through multiregion and longitudinal sampling, evolutionary processes have been detailed in the tumor and the immune microenvironment in non-small cell lung cancer and clear-cell renal cell carcinoma. TRACERx has revealed the potential therapeutic utility of targeting clonal neoantigens and ctDNA detection in the adjuvant setting as a minimal residual disease detection tool primed for translation into clinical trials.
©2021 American Association for Cancer Research.
Conflict of interest statement
CS is Royal Society Napier Research Professor. This work was supported by the Francis Crick Institute that receives its core funding from Cancer Research UK (FC001169), the UK Medical Research Council (FC001169), and the Wellcome Trust (FC001169). CS is funded by Cancer Research UK (TRACERx, PEACE and CRUK Cancer Immunotherapy Catalyst Network), Cancer Research UK Lung Cancer Centre of Excellence, the Rosetrees Trust, Butterfield and Stoneygate Trusts, NovoNordisk Foundation (ID16584), Royal Society Professorship Enhancement Award (RP/EA/180007), the NIHR BRC at University College London Hospitals, and the CRUK-UCL Centre, Experimental Cancer Medicine Centre, and the Breast Cancer Research Foundation (BCRF). This research is supported by a Stand Up To Cancer‐LUNGevity-American Lung Association Lung Cancer Interception Dream Team Translational Research Grant (Grant Number: SU2C-AACR-DT23-17). Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the Scientific Partner of SU2C. CS receives funding from the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7/2007-2013) Consolidator Grant (FP7-THESEUS-617844), European Commission ITN (FP7-PloidyNet 607722), an ERC Advanced Grant (PROTEUS) from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 835297), and Chromavision from the European Union’s Horizon 2020 research and innovation programme (grant agreement 665233). CS acknowledges grant support from Pfizer, AstraZeneca, Bristol Myers Squibb, Roche-Ventana, Boehringer-Ingelheim, Archer Dx Inc (collaboration in minimal residual disease sequencing technologies) and Ono Pharmaceutical, is an AstraZeneca Advisory Board member and Chief Investigator for the MeRmaiD1 clinical trial, has consulted for Pfizer, Novartis, GlaxoSmithKline, MSD, Bristol Myers Squibb, Celgene, AstraZeneca, Illumina, Amgen, Genentech, Roche-Ventana, GRAIL, Medicxi, Bicycle Therapeutics, and the Sarah Cannon Research Institute, has stock options in Apogen Biotechnologies, Epic Bioscience, GRAIL, and has stock options and is co-founder of Achilles Therapeutics. CS holds European patents relating to assay technology to detect tumour recurrence (PCT/GB2017/053289); to targeting neoantigens (PCT/EP2016/059401), identifying patent response to immune checkpoint blockade (PCT/EP2016/071471), determining HLA LOH (PCT/GB2018/052004), predicting survival rates of patients with cancer (PCT/GB2020/050221), identifying patients who respond to cancer treatment (PCT/GB2018/051912), a US patent relating to detecting tumour mutations (PCT/US2017/28013) and both a European and US patent related to identifying insertion/deletion mutation targets (PCT/GB2018/051892).
ST is funded by Cancer Research UK (grant reference number C50947/A18176), the National Institute for Health Research (NIHR) Biomedical Research Centre at the Royal Marsden Hospital and Institute of Cancer Research (grant reference number A109), the Kidney and Melanoma Cancer Fund of The Royal Marsden Cancer Charity, The Rosetrees Trust (grant reference number A2204), Ventana Medical Systems Inc (grant reference numbers 10467 and 10530), the National Institute of Health U01 Award and Melanoma Research Alliance. ST has received speaking fees from Roche, Astra Zeneca, Novartis and Ipsen. ST has the following patents filed: Indel mutations as a therapeutic target and predictive biomarker PCTGB2018/051892 and PCTGB2018/051893 and Clear Cell Renal Cell Carcinoma Biomarkers P113326GB.
MJH has received funding from Cancer Research UK, National Institute for Health Research, Rosetrees Trust, UKI NETs and NIHR University College London Hospitals Biomedical Research Centre. MJH is a member of the Scientific Advisory Board and Steering Committee for Achilles Therapeutics.
Figures

References
-
- Turajlic S, Sottoriva A, Graham T, Swanton C. Resolving genetic heterogeneity in cancer. Nat Rev Genet. 2019;20:404–16. [Internet]. Available from: http://www.nature.com/articles/s41576-019-0114-6. - PubMed
-
- Turajlic S, Xu H, Litchfield K, Rowan A, Horswell S, Chambers T, et al. Deterministic Evolutionary Trajectories Influence Primary Tumor Growth: TRACERx Renal. Cell. 2018;173:595–610.:e11. [Internet]. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867418303751. - PMC - PubMed
-
- McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis GM, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. [Internet]. Available from: http://www.nature.com/articles/nature07385. - PMC - PubMed
-
- Campbell PJ, Getz G, Korbel JO, Stuart JM, Jennings JL, Stein LD, et al. Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. [Internet]. Available from: http://www.nature.com/articles/s41586-020-1969-6. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- 617844/ERC_/European Research Council/International
- 835297/ERC_/European Research Council/International
- 24956/CRUK_/Cancer Research UK/United Kingdom
- 20466/CRUK_/Cancer Research UK/United Kingdom
- 25253/CRUK_/Cancer Research UK/United Kingdom
- 17786/CRUK_/Cancer Research UK/United Kingdom
- 30025/CRUK_/Cancer Research UK/United Kingdom
- 29911/CRUK_/Cancer Research UK/United Kingdom
- 218274/WT_/Wellcome Trust/United Kingdom
- MR/P014712/1/MRC_/Medical Research Council/United Kingdom
- 211179/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- FC001169/WT_/Wellcome Trust/United Kingdom
- A18176/CRUK_/Cancer Research UK/United Kingdom
- MR/V033077/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

