Deep generative models in DataSHIELD
- PMID: 33812380
- PMCID: PMC8019187
- DOI: 10.1186/s12874-021-01237-6
Deep generative models in DataSHIELD
Abstract
Background: The best way to calculate statistics from medical data is to use the data of individual patients. In some settings, this data is difficult to obtain due to privacy restrictions. In Germany, for example, it is not possible to pool routine data from different hospitals for research purposes without the consent of the patients.
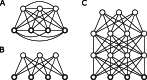
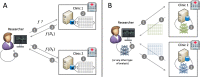
Methods: The DataSHIELD software provides an infrastructure and a set of statistical methods for joint, privacy-preserving analyses of distributed data. The contained algorithms are reformulated to work with aggregated data from the participating sites instead of the individual data. If a desired algorithm is not implemented in DataSHIELD or cannot be reformulated in such a way, using artificial data is an alternative. Generating artificial data is possible using so-called generative models, which are able to capture the distribution of given data. Here, we employ deep Boltzmann machines (DBMs) as generative models. For the implementation, we use the package "BoltzmannMachines" from the Julia programming language and wrap it for use with DataSHIELD, which is based on R.
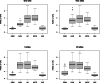
Results: We present a methodology together with a software implementation that builds on DataSHIELD to create artificial data that preserve complex patterns from distributed individual patient data. Such data sets of artificial patients, which are not linked to real patients, can then be used for joint analyses. As an exemplary application, we conduct a distributed analysis with DBMs on a synthetic data set, which simulates genetic variant data. Patterns from the original data can be recovered in the artificial data using hierarchical clustering of the virtual patients, demonstrating the feasibility of the approach. Additionally, we compare DBMs, variational autoencoders, generative adversarial networks, and multivariate imputation as generative approaches by assessing the utility and disclosure of synthetic data generated from real genetic variant data in a distributed setting with data of a small sample size.
Conclusions: Our implementation adds to DataSHIELD the ability to generate artificial data that can be used for various analyses, e.g., for pattern recognition with deep learning. This also demonstrates more generally how DataSHIELD can be flexibly extended with advanced algorithms from languages other than R.
Keywords: Biomedical research/methods; Deep learning; Distributed system; Privacy/statistics and numerical data.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
-
- Nowok B, Raab GM, Dibben C. Synthpop: bespoke creation of synthetic data in R. J Stat Softw. 2016;74:1–26. doi: 10.18637/jss.v074.i11. - DOI
-
- Manrique-Vallier D, Hu J. Bayesian non-parametric generation of fully synthetic multivariate categorical data in the presence of structural zeros. J R Stat Soc Ser A Stat Soc. 2018;181:635–647. doi: 10.1111/rssa.12352. - DOI
-
- Quick H, Holan SH, Wikle CK. Generating partially synthetic geocoded public use data with decreased disclosure risk by using differential smoothing. J R Stat Soc Ser A Stat Soc. 2018;181:649–661. doi: 10.1111/rssa.12360. - DOI
-
- Statice GmbH. Company web site. https://www.statice.ai/. Accessed 27 Aug 2019.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

