Sticky problems: extraction of nucleic acids from molluscs
- PMID: 33813891
- PMCID: PMC8059567
- DOI: 10.1098/rstb.2020.0162
Sticky problems: extraction of nucleic acids from molluscs
Abstract
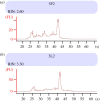
Traditional molecular methods and omics-techniques across molluscan taxonomy increasingly inform biology of Mollusca. Recovery of DNA and RNA for such studies is challenged by common biological properties of the highly diverse molluscs. Molluscan biomineralization, adhesive structures and mucus involve polyphenolic proteins and mucopolysaccharides that hinder DNA extraction or copurify to inhibit enzyme-catalysed molecular procedures. DNA extraction methods that employ the detergent hexadecyltrimethylammoniumbromide (CTAB) to remove these contaminants importantly facilitate molecular-level study of molluscs. Molluscan pigments may stain DNA samples and interfere with spectrophotometry, necessitating gel electrophoresis or fluorometry for accurate quantification. RNA can reliably be extracted but the 'hidden break' in 28S rRNA of molluscs (like most protostomes) causes 18S and 28S rRNA fragments to co-migrate electrophoretically. This challenges the standard quality control based on the ratio of 18S and 28S rRNA, developed for deuterostome animals. High-AT content in molluscan rRNA prevents the effective purification of polyadenylated mRNA. Awareness of these matters aids the continuous expansion of molecular malacology, enabling work also with museum specimens and next-generation sequencing, with the latter imposing unprecedented demands on DNA quality. Alternative methods to extract nucleic acids from molluscs are available from literature and, importantly, from communications with others who study the molecular biology of molluscs. This article is part of the Theo Murphy meeting issue 'Molluscan genomics: broad insights and future directions for a neglected phylum'.
Keywords: AT rich; CTAB; hidden break; mollusca; mucopolysaccharides; pigment.
Figures

References
-
- Haszprunar G, Schander C, Halanych KM. 2008. Relationships of higher molluscan taxa. In Phylogeny and evolution of the mollusca (eds Ponder W, Lindberg DR), pp. 19-32. Berkeley, CA: University of California Press.
-
- Maniatis T, Fritsch EF, Sambrook J. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

