Ribosome Rescue Pathways in Bacteria
- PMID: 33815344
- PMCID: PMC8012679
- DOI: 10.3389/fmicb.2021.652980
Ribosome Rescue Pathways in Bacteria
Abstract
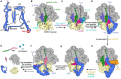
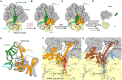
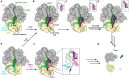
Ribosomes that become stalled on truncated or damaged mRNAs during protein synthesis must be rescued for the cell to survive. Bacteria have evolved a diverse array of rescue pathways to remove the stalled ribosomes from the aberrant mRNA and return them to the free pool of actively translating ribosomes. In addition, some of these pathways target the damaged mRNA and the incomplete nascent polypeptide chain for degradation. This review highlights the recent developments in our mechanistic understanding of bacterial ribosomal rescue systems, including drop-off, trans-translation mediated by transfer-messenger RNA and small protein B, ribosome rescue by the alternative rescue factors ArfA and ArfB, as well as Bacillus ribosome rescue factor A, an additional rescue system found in some Gram-positive bacteria, such as Bacillus subtilis. Finally, we discuss the recent findings of ribosome-associated quality control in particular bacterial lineages mediated by RqcH and RqcP. The importance of rescue pathways for bacterial survival suggests they may represent novel targets for the development of new antimicrobial agents against multi-drug resistant pathogenic bacteria.
Keywords: ArfA; ArfB; RqcH; SmpB; peptidyl-tRNA drop-off; ribosome rescue; ribosome-associated quality control; tmRNA.
Copyright © 2021 Müller, Crowe-McAuliffe and Wilson.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

