Interdependent Polar Localization of FlhF and FlhG and Their Importance for Flagellum Formation of Vibrio parahaemolyticus
- PMID: 33815347
- PMCID: PMC8009987
- DOI: 10.3389/fmicb.2021.655239
Interdependent Polar Localization of FlhF and FlhG and Their Importance for Flagellum Formation of Vibrio parahaemolyticus
Abstract
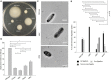
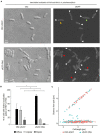
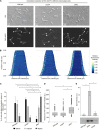
Failure of the cell to properly regulate the number and intracellular positioning of their flagella, has detrimental effects on the cells' swimming ability. The flagellation pattern of numerous bacteria is regulated by the NTPases FlhF and FlhG. In general, FlhG controls the number of flagella produced, whereas FlhF coordinates the position of the flagella. In the human pathogen Vibrio parahaemolyticus, its single flagellum is positioned and formed at the old cell pole. Here, we describe the spatiotemporal localization of FlhF and FlhG in V. parahaemolyticus and their effect on swimming motility. Absence of either FlhF or FlhG caused a significant defect in swimming ability, resulting in absence of flagella in a ΔflhF mutant and an aberrant flagellated phenotype in ΔflhG. Both proteins localized to the cell pole in a cell cycle-dependent manner, but displayed different patterns of localization throughout the cell cycle. FlhF transitioned from a uni- to bi-polar localization, as observed in other polarly flagellated bacteria. Localization of FlhG was strictly dependent on the cell pole-determinant HubP, while polar localization of FlhF was HubP independent. Furthermore, localization of FlhF and FlhG was interdependent and required for each other's proper intracellular localization and recruitment to the cell pole. In the absence of HubP or FlhF, FlhG forms non-polar foci in the cytoplasm of the cell, suggesting the possibility of a secondary localization site within the cell besides its recruitment to the cell poles.
Keywords: FlhF; FlhG; HubP; Vibrio parahaemolyticus; flagellum; intracellular organization.
Copyright © 2021 Arroyo-Pérez and Ringgaard.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

