Initial Insights Into the Genetic Epidemiology of SARS-CoV-2 Isolates From Kerala Suggest Local Spread From Limited Introductions
- PMID: 33815467
- PMCID: PMC8010186
- DOI: 10.3389/fgene.2021.630542
Initial Insights Into the Genetic Epidemiology of SARS-CoV-2 Isolates From Kerala Suggest Local Spread From Limited Introductions
Abstract
Coronavirus disease 2019 (COVID-19) rapidly spread from a city in China to almost every country in the world, affecting millions of individuals. The rapid increase in the COVID-19 cases in the state of Kerala in India has necessitated the understanding of SARS-CoV-2 genetic epidemiology. We sequenced 200 samples from patients in Kerala using COVIDSeq protocol amplicon-based sequencing. The analysis identified 166 high-quality single-nucleotide variants encompassing four novel variants and 89 new variants in the Indian isolated SARS-CoV-2. Phylogenetic and haplotype analysis revealed that the virus was dominated by three distinct introductions followed by local spread suggesting recent outbreaks and that it belongs to the A2a clade. Further analysis of the functional variants revealed that two variants in the S gene associated with increased infectivity and five variants mapped in primer binding sites affect the efficacy of RT-PCR. To the best of our knowledge, this is the first and most comprehensive report of SARS-CoV-2 genetic epidemiology from Kerala.
Keywords: COVID-19; COVIDSeq; Kerala; genetic epidemiology; variants.
Copyright © 2021 Radhakrishnan, Divakar, Jain, Viswanathan, Bhoyar, Jolly, Imran, Sharma, Rophina, Ranjan, Sehgal, Jose, Raman, Kesavan, George, Mathew, Poovullathil, Keeriyatt Govindan, Nair, Vadekkandiyil, Gladson, Mohan, Parambath, Mangla, Shamnath, Indian CoV2 Genomics & Genetic Epidemiology (IndiCovGEN) Consortium, Sivasubbu and Scaria.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
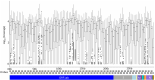
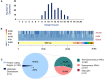


References
-
- Bedford T. (2020). Phylodynamic Estimation of Incidence and Prevalence of Novel Coronavirus (nCoV) Infections Through Time. Virological. Available online at: http://virological.org/t/phylodynamic-estimation-of-incidence-and-preval... (accessed August 30, 2020).
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

