Staphylococcus aureus membrane vesicles contain immunostimulatory DNA, RNA and peptidoglycan that activate innate immune receptors and induce autophagy
- PMID: 33815695
- PMCID: PMC8015888
- DOI: 10.1002/jev2.12080
Staphylococcus aureus membrane vesicles contain immunostimulatory DNA, RNA and peptidoglycan that activate innate immune receptors and induce autophagy
Abstract
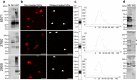
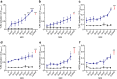
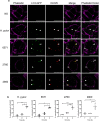
Gram-positive bacteria ubiquitously produce membrane vesicles (MVs), and although they contribute to biological functions, our knowledge regarding their composition and immunogenicity remains limited. Here we examine the morphology, contents and immunostimulatory functions of MVs produced by three Staphylococcus aureus strains; a methicillin resistant clinical isolate, a methicillin sensitive clinical isolate and a laboratory-adapted strain. We observed differences in the number and morphology of MVs produced by each strain and showed that they contain microbe-associated molecular patterns (MAMPs) including protein, nucleic acids and peptidoglycan. Analysis of MV-derived RNA indicated the presence of small RNA (sRNA). Furthermore, we detected variability in the amount and composition of protein, nucleic acid and peptidoglycan cargo carried by MVs from each S. aureus strain. S. aureus MVs activated Toll-like receptor (TLR) 2, 7, 8, 9 and nucleotide-binding oligomerization domain containing protein 2 (NOD2) signalling and promoted cytokine and chemokine release by epithelial cells, thus identifying that MV-associated MAMPs including DNA, RNA and peptidoglycan are detected by pattern recognition receptors (PRRs). Moreover, S. aureus MVs induced the formation of and colocalized with autophagosomes in epithelial cells, while inhibition of lysosomal acidification using bafilomycin A1 resulted in accumulation of autophagosomal puncta that colocalized with MVs, revealing the ability of the host to degrade MVs via autophagy. This study reveals the ability of DNA, RNA and peptidoglycan associated with MVs to activate PRRs in host epithelial cells, and their intracellular degradation via autophagy. These findings advance our understanding of the immunostimulatory roles of Gram-positive bacterial MVs in mediating pathogenesis, and their intracellular fate within the host.
Keywords: DNA; NOD2; RNA; Staphylococcus aureus; TLRs; autophagy; bacterial membrane vesicles; bacterial pathogenesis; innate immunity; peptidoglycan.
© 2021 The Authors. Journal of Extracellular Vesicles published by Wiley Periodicals, LLC on behalf of the International Society for Extracellular Vesicles.
Figures

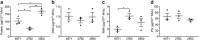



References
-
- Andreoni, F. , Toyofuku, M. , Menzi, C. , Kalawong, R. , Mairpady Shambat, S. , François, P. , Zinkernagel, A. S. , & Eberl, L. (2019). Antibiotics stimulate formation of vesicles in Staphylococcus aureus in both phage‐dependent and ‐independent fashions and via different routes. Antimicrobial Agents and Chemotherapy, 63, e01439–01418. - PMC - PubMed
-
- Avila‐Calderón, E. D. , Lopez‐Merino, A. , Jain, N. , Peralta, H. , López‐Villegas, E. O. , Sriranganathan, N. , Boyle, S. M. , Witonsky, S. , & Contreras‐Rodríguez, A. (2012). Characterization of outer membrane vesicles from Brucella melitensis and protection induced in mice. Clinical & Developmental Immunology, 2012, 352493. - PMC - PubMed
-
- Bielaszewska, M. , Rüter, C. , Bauwens, A. , Greune, L. , Jarosch, K.‐A. , Steil, D. , Zhang, W. , He, X. , Lloubes, R. , Fruth, A. , Kim, K. S. , Schmidt, M. A. , Dobrindt, U. , Mellmann, A. , & Karch, H. (2017). Host cell interactions of outer membrane vesicle‐associated virulence factors of enterohemorrhagic Escherichia coli O157: Intracellular delivery, trafficking and mechanisms of cell injury. Plos Pathogens, 13, e1006159. - PMC - PubMed
-
- Bielig, H. , Rompikuntal, P. K. , Dongre, M. , Zurek, B. , Lindmark, B. , Ramstedt, M. , Wai, S. N. , & Kufer, T. A. (2011). NOD‐like receptor activation by outer membrane vesicles from Vibrio cholerae non‐O1 non‐O139 strains is modulated by the quorum‐sensing regulator HapR. Infection and Immunity, 79, 1418–1427. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

