Nilotinib Improves Bioenergetic Profiling in Brain Astroglia in the 3xTg Mouse Model of Alzheimer's Disease
- PMID: 33815876
- PMCID: PMC7990369
- DOI: 10.14336/AD.2020.0910
Nilotinib Improves Bioenergetic Profiling in Brain Astroglia in the 3xTg Mouse Model of Alzheimer's Disease
Abstract
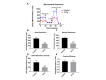
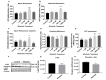
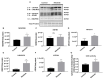
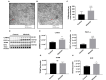
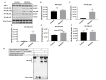
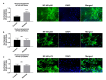
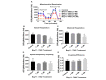
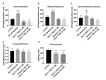
Current treatments targeting amyloid beta in Alzheimer's disease (AD) have minimal efficacy, which results in a huge unmet medical need worldwide. Accumulating data suggest that brain mitochondrial dysfunction play a critical role in AD pathogenesis. Targeting cellular mechanisms associated with mitochondrial dysfunction in AD create a novel approach for drug development. This study investigated the effects of nilotinib, as a selective tyrosine kinase inhibitor, in astroglia derived from 3xTg-AD mice versus their C57BL/6-controls. Parameters included oxygen consumption rates (OCR), ATP, cytochrome c oxidase (COX), citrate synthase (CS) activity, alterations in oxidative phosphorylation (OXPHOS), nuclear factor kappa B (NF-κB), key regulators of mitochondrial dynamics (mitofusin (Mfn1), dynamin-related protein 1 (Drp1)), and mitochondrial biogenesis (peroxisome proliferator-activated receptor gamma coactivator1-alpha (PGC-1α), calcium/calmodulin-dependent protein kinase II (CaMKII), and nuclear factor (erythroid-derived 2)-like 2 (Nrf2)). Nilotinib increased OCR, ATP, COX, Mfn1, and OXPHOS levels in 3xTg astroglia. No significant differences were detected in levels of Drp1 protein and CS activity. Nilotinib enhanced mitochondrial numbers, potentially through a CaMKII-PGC1α-Nrf2 pathway in 3xTg astroglia. Additionally, nilotinib-induced OCR increases were reduced in the presence of the NF-κB inhibitor, Bay11-7082. The data suggest that NF-κB signaling is intimately involved in nilotinib-induced changes in bioenergetics in 3xTg brain astroglia. Nilotinib increased translocation of the NF-κB p50 subunit into the nucleus of 3xTg astroglia that correlates with an increased expression and activation of NF-κB. The current findings support a role for nilotinib in improving mitochondrial function and suggest that astroglia may be a key therapeutic target in treating AD.
Keywords: Alzheimer’s disease; astroglia; bioenergetics; biogenesis; citrate synthase; cytochrome c oxidase; mitochondrial function; nuclear factor kappa B (NF-κB); oxidative phosphorylation.
copyright: © 2021 Adlimoghaddam et al.
Conflict of interest statement
Conflict of Interest The authors declare no conflict of interest.
Figures




References
-
- McKeith I, Cummings J (2005). Behavioural changes and psychological symptoms in dementia disorders. Lancet Neurol, 4:735-742. - PubMed
-
- Glabe CC (2005). Amyloid accumulation and pathogensis of Alzheimer's disease: significance of monomeric, oligomeric and fibrillar Abeta. Subcell Biochem, 38:167-177. - PubMed
-
- Hardy J, Selkoe DJ (2002). The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science, 297:353-356. - PubMed
-
- Cardoso S, Seica RM, Moreira PI (2017). Mitochondria as a target for neuroprotection: implications for Alzheimer s disease. Expert Rev Neurother, 17:77-91. - PubMed
-
- Cadonic C, Sabbir MG, Albensi BC (2016). Mechanisms of Mitochondrial Dysfunction in Alzheimer's Disease. Mol Neurobiol, 53:6078-6090. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
