Genomic Characterization of Campylobacter jejuni Adapted to the Guinea Pig (Cavia porcellus) Host
- PMID: 33816330
- PMCID: PMC8012767
- DOI: 10.3389/fcimb.2021.607747
Genomic Characterization of Campylobacter jejuni Adapted to the Guinea Pig (Cavia porcellus) Host
Abstract
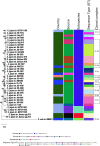
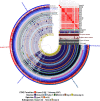
Campylobacter jejuni is the leading bacterial cause of gastroenteritis worldwide with excessive incidence in low-and middle-income countries (LMIC). During a survey for C. jejuni from putative animal hosts in a town in the Peruvian Amazon, we were able to isolate and whole genome sequence two C. jejuni strains from domesticated guinea pigs (Cavia porcellus). The C. jejuni isolated from guinea pigs had a novel multilocus sequence type that shared some alleles with other C. jejuni collected from guinea pigs. Average nucleotide identity and phylogenetic analysis with a collection of C. jejuni subsp. jejuni and C. jejuni subsp. doylei suggest that the guinea pig isolates are distinct. Genomic comparisons demonstrated gene gain and loss that could be associated with guinea pig host specialization related to guinea pig diet, anatomy, and physiology including the deletion of genes involved with selenium metabolism, including genes encoding the selenocysteine insertion machinery and selenocysteine-containing proteins.
Keywords: Campylobacter jejuni; campylobacteriosis; gastroenteritis; selenocysteine; source attribution.
Copyright © 2021 Parker, Cooper, Schiaffino, Miller, Huynh, Gray, Olortegui, Bardales, Trigoso, Penataro-Yori and Kosek.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Atterby C., Mourkas E., Meric G., Pascoe B., Wang H., Waldenstrom J., et al. . (2018). The Potential of Isolation Source to Predict Colonization in Avian Hosts: A Case Study in Campylobacter jejuni Strains From Three Bird Species. Front. Microbiol. 9, 591. 10.3389/fmicb.2018.00591 - DOI - PMC - PubMed
-
- Burakoff A., Brown K., Knutsen J., Hopewell C., Rowe S., Bennett C., et al. . (2018). Outbreak of Fluoroquinolone-Resistant Campylobacter jejuni Infections Associated with Raw Milk Consumption from a Herdshare Dairy - Colorado, 2016. MMWR Morb. Mortal. Wkly. Rep. 67, 146–148. 10.15585/mmwr.mm6705a2 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

