Taxon-Specific Proteins of the Pathogenic Entamoeba Species E. histolytica and E. nuttalli
- PMID: 33816346
- PMCID: PMC8017271
- DOI: 10.3389/fcimb.2021.641472
Taxon-Specific Proteins of the Pathogenic Entamoeba Species E. histolytica and E. nuttalli
Abstract
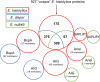
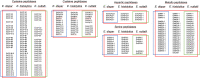
The human protozoan parasite Entamoeba histolytica can live in the human intestine for months or years without generating any symptoms in the host. For unknown reasons, amoebae can suddenly destroy the intestinal mucosa and become invasive. This can lead to amoebic colitis or extraintestinal amoebiasis whereby the amoebae spread to other organs via the blood vessels, most commonly the liver where abscesses develop. Entamoeba nuttalli is the closest genetic relative of E. histolytica and is found in wild macaques. Another close relative is E. dispar, which asyptomatically infects the human intestine. Although all three species are closely related, only E. histolytica and E. nuttalli are able to penetrate their host's intestinal epithelium. Lineage-specific genes and gene families may hold the key to understanding differences in virulence among species. Here we discuss those genes found in E. histolytica that have relatives in only one or neither of its sister species, with particular focus on the peptidase, AIG, Ariel, and BspA families.
Keywords: AIG; Ariel; BspA; Entamoeba; peptidases; virulence.
Copyright © 2021 König, Honecker, Wilson, Weedall, Hall, Roeder, Metwally and Bruchhaus.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Human amebiasis: breaking the paradigm?Int J Environ Res Public Health. 2010 Mar;7(3):1105-20. doi: 10.3390/ijerph7031105. Epub 2010 Mar 16. Int J Environ Res Public Health. 2010. PMID: 20617021 Free PMC article.
-
Proteomic comparison of Entamoeba histolytica and Entamoeba dispar and the role of E. histolytica alcohol dehydrogenase 3 in virulence.PLoS Negl Trop Dis. 2009;3(4):e415. doi: 10.1371/journal.pntd.0000415. Epub 2009 Apr 14. PLoS Negl Trop Dis. 2009. PMID: 19365541 Free PMC article.
-
Intestinal amoebiasis: 160 years of its first detection and still remains as a health problem in developing countries.Int J Med Microbiol. 2020 Jan;310(1):151358. doi: 10.1016/j.ijmm.2019.151358. Epub 2019 Sep 19. Int J Med Microbiol. 2020. PMID: 31587966 Review.
-
Whole genome sequencing of Entamoeba nuttalli reveals mammalian host-related molecular signatures and a novel octapeptide-repeat surface protein.PLoS Negl Trop Dis. 2019 Dec 5;13(12):e0007923. doi: 10.1371/journal.pntd.0007923. eCollection 2019 Dec. PLoS Negl Trop Dis. 2019. PMID: 31805050 Free PMC article.
-
Entamoeba histolytica and Entamoeba dispar are distinct species; clinical, epidemiological and serological evidence.Int J Parasitol. 1998 Jan;28(1):181-6. doi: 10.1016/s0020-7519(97)00177-x. Int J Parasitol. 1998. PMID: 9504344 Review.
Cited by
-
Gene expression of axenically-isolated clinical Entamoeba histolytica strains and its impact on disease severity of amebiasis.PLoS Pathog. 2022 Sep 30;18(9):e1010880. doi: 10.1371/journal.ppat.1010880. eCollection 2022 Sep. PLoS Pathog. 2022. PMID: 36178974 Free PMC article.
-
A landscape of gene regulation in the parasitic amoebozoa Entamoeba spp.PLoS One. 2022 Aug 1;17(8):e0271640. doi: 10.1371/journal.pone.0271640. eCollection 2022. PLoS One. 2022. PMID: 35913975 Free PMC article.
-
Genes differentially expressed between pathogenic and non-pathogenic Entamoeba histolytica clones influence pathogenicity-associated phenotypes by multiple mechanisms.PLoS Pathog. 2023 Dec 22;19(12):e1011745. doi: 10.1371/journal.ppat.1011745. eCollection 2023 Dec. PLoS Pathog. 2023. PMID: 38134215 Free PMC article.
-
Attenuation of In Vitro and In Vivo Virulence Is Associated with Repression of Gene Expression of AIG1 Gene in Entamoeba histolytica.Pathogens. 2023 Mar 21;12(3):489. doi: 10.3390/pathogens12030489. Pathogens. 2023. PMID: 36986411 Free PMC article.
-
Amoebiasis: Advances in Diagnosis, Treatment, Immunology Features and the Interaction with the Intestinal Ecosystem.Int J Mol Sci. 2023 Jul 21;24(14):11755. doi: 10.3390/ijms241411755. Int J Mol Sci. 2023. PMID: 37511519 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

