Deciphering Epitranscriptome: Modification of mRNA Bases Provides a New Perspective for Post-transcriptional Regulation of Gene Expression
- PMID: 33816473
- PMCID: PMC8010680
- DOI: 10.3389/fcell.2021.628415
Deciphering Epitranscriptome: Modification of mRNA Bases Provides a New Perspective for Post-transcriptional Regulation of Gene Expression
Abstract
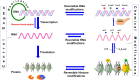
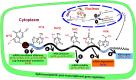
Gene regulation depends on dynamic and reversibly modifiable biological and chemical information in the epigenome/epitranscriptome. Accumulating evidence suggests that messenger RNAs (mRNAs) are generated in flashing bursts in the cells in a precisely regulated manner. However, the different aspects of the underlying mechanisms are not fully understood. Cellular RNAs are post-transcriptionally modified at the base level, which alters the metabolism of mRNA. The current understanding of epitranscriptome in the animal system is far ahead of that in plants. The accumulating evidence indicates that the epitranscriptomic changes play vital roles in developmental processes and stress responses. Besides being non-genetically encoded, they can be of reversible nature and involved in fine-tuning the expression of gene. However, different aspects of base modifications in mRNAs are far from adequate to assign the molecular basis/functions to the epitranscriptomic changes. Advances in the chemogenetic RNA-labeling and high-throughput next-generation sequencing techniques are enabling functional analysis of the epitranscriptomic modifications to reveal their roles in mRNA biology. Mapping of the common mRNA modifications, including N 6-methyladenosine (m6A), and 5-methylcytidine (m5C), have enabled the identification of other types of modifications, such as N 1-methyladenosine. Methylation of bases in a transcript dynamically regulates the processing, cellular export, translation, and stability of the mRNA; thereby influence the important biological and physiological processes. Here, we summarize the findings in the field of mRNA base modifications with special emphasis on m6A, m5C, and their roles in growth, development, and stress tolerance, which provide a new perspective for the regulation of gene expression through post-transcriptional modification. This review also addresses some of the scientific and technical issues in epitranscriptomic study, put forward the viewpoints to resolve the issues, and discusses the future perspectives of the research in this area.
Keywords: 5-methylcytidine; N6-methyladenosine; RNA metabolism; RNA modification; central dogma; epitranscriptomics; mRNA methylation; post-transcriptional regulation.
Copyright © 2021 Kumar and Mohapatra.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

