The lncRNA DLGAP1-AS1/miR-149-5p/TGFB2 axis contributes to colorectal cancer progression and 5-FU resistance by regulating smad2 pathway
- PMID: 33816780
- PMCID: PMC7985718
- DOI: 10.1016/j.omto.2021.01.003
The lncRNA DLGAP1-AS1/miR-149-5p/TGFB2 axis contributes to colorectal cancer progression and 5-FU resistance by regulating smad2 pathway
Abstract
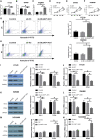
Colorectal carcinoma (CRC) ranks as the third most common malignancy. Long non-coding RNA DLGAP1-AS1 was reported to be dysregulated and to play a pivotal role in hepatocellular carcinoma (HCC). This work aims to analyze the functions and molecular basis of DLGAP1-AS1 in CRC progression and 5-fluorouracil resistance. Cell Counting Kit-8 (CCK-8) assay, Transwell assay, flow cytometry, and western blot were utilized to measure the CRC cell activity, invasiveness, and apoptosis. RNA immunoprecipitation (RIP) and dual-luciferase reporter gene assay were adopted to verify the direct mutual action between DLGAP1-AS1 and miR-149-5p. The effect of DLGAP1-AS1 knockdown on tumor growth and chemosensitivity of 5-fluorouracil (5-FU) were investigated in the mouse CRC xenograft models. Functional assays showed that silencing DLGAP1-AS1 expression remarkably inhibited cell proliferation and aggressiveness ability and enhanced apoptosis rate and cell chemosensitivity to 5-FU. In addition, miR-149-5p was identified as a tumor suppressor and a direct downstream target of DLGAP1-AS1 in CRC. Furthermore, miR-149-5p was confirmed to directly bind to TGFB2 and DLGAP1-AS1 could regulate the expression of TGFB2 signaling pathway via miR-149-5p in CRC. These new findings indicate that DLGAP1-AS1 knockdown inhibited the progression of CRC and enhanced the 5-FU sensitivity of CRC cells through miR-149-5p/TGFB2 regulatory axis, suggesting that DLGAP1-AS1 may be a promising therapeutic target for CRC.
Keywords: DLGAP1-AS1; TGFB2; chemoresistance; colorectal cancer; lncRNA; miR-149-5p.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
-
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018;68:7–30. - PubMed
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. - PubMed
-
- Zhong X., Fang Y.J., Pan Z.Z., Lu M.S., Zheng M.C., Chen Y.M., Zhang C.X. Dietary fiber and fiber fraction intakes and colorectal cancer risk in Chinese adults. Nutr. Cancer. 2014;66:351–361. - PubMed
-
- Torre L.A., Siegel R.L., Ward E.M., Jemal A. Global Cancer Incidence and Mortality Rates and Trends--An Update. Cancer Epidemiol. Biomarkers Prev. 2016;25:16–27. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

