Database limitations for studying the human gut microbiome
- PMID: 33816940
- PMCID: PMC7924478
- DOI: 10.7717/peerj-cs.289
Database limitations for studying the human gut microbiome
Abstract
Background: In the last twenty years, new methodologies have made possible the gathering of large amounts of data concerning the genetic information and metabolic functions associated to the human gut microbiome. In spite of that, processing all this data available might not be the simplest of tasks, which could result in an excess of information awaiting proper annotation. This assessment intended on evaluating how well respected databases could describe a mock human gut microbiome.
Methods: In this work, we critically evaluate the output of the cross-reference between the Uniprot Knowledge Base (Uniprot KB) and the Kyoto Encyclopedia of Genes and Genomes Orthologs (KEGG Orthologs) or the evolutionary genealogy of genes: Non-supervised Orthologous groups (EggNOG) databases regarding a list of species that were previously found in the human gut microbiome.
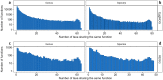
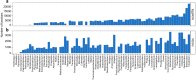
Results: From a list which contemplates 131 species and 52 genera, 53 species and 40 genera had corresponding entries for KEGG Database and 82 species and 47 genera had corresponding entries for EggNOG Database. Moreover, we present the KEGG Orthologs (KOs) and EggNOG Orthologs (NOGs) entries associated to the search as their distribution over species and genera and lists of functions that appeared in many species or genera, the "core" functions of the human gut microbiome. We also present the relative abundance of KOs and NOGs throughout phyla and genera. Lastly, we expose a variance found between searches with different arguments on the database entries. Inferring functionality based on cross-referencing UniProt and KEGG or EggNOG can be lackluster due to the low number of annotated species in Uniprot and due to the lower number of functions affiliated to the majority of these species. Additionally, the EggNOG database showed greater performance for a cross-search with Uniprot about a mock human gut microbiome. Notwithstanding, efforts targeting cultivation, single-cell sequencing or the reconstruction of high-quality metagenome-assembled genomes (MAG) and their annotation are needed to allow the use of these databases for inferring functionality in human gut microbiome studies.
Keywords: Database; EggNOG; Functional diversity; Gut microbiome; Human Microbiome; KEGG; Uniprot.
©2020 Dias et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


References
-
- Chauhan NS, Pandey R, Mondal AK, Gupta S, Verma MK, Jain S, Ahmed V, Patil R, Agarwal D, Girase B, Shrivastava A. Western Indian rural gut microbial diversity in extreme Prakriti endo-phenotypes reveals signature microbes. Frontiers in Microbiology. 2018;9(118):1–12. doi: 10.3389/fmicb.2018.00118. - DOI - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Research Materials
