Temporal oral microbiome changes with brushing in children with cleft lip and palate
- PMID: 33817376
- PMCID: PMC8005767
- DOI: 10.1016/j.heliyon.2021.e06513
Temporal oral microbiome changes with brushing in children with cleft lip and palate
Abstract
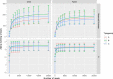
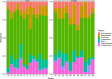
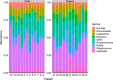
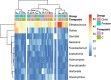
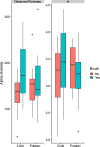
This cohort study aimed to characterize the oral microbiome of children with CLP, from two different age groups, and evaluate the effect of supervised or unsupervised toothbrushing on the microbiome of the cleft over time. Swab samples were collected from the cleft area at three different time points (A; no brushing, B; after 15 days and C; after 30 days) and were analyzed using next-generation sequencing to determine the microbial composition and diversity in these time points. Overall, brushing significantly decreased the abundance of the genera Alloprevotella and Leptotrichia in the two age groups examined, and for Alloprevotella this decrease was more evident for children (2-6 years old). In the preteen group (7-12 years old), a significant relative increase of the genus Rothia was observed after brushing. In this study, the systematic brushing over a period of thirty days also resulted in differences at the intra-individual bacterial richness.
Keywords: Alloprevotella; CLP; Children and preteen; Leptotrichia; Microbiome; Next-generation sequencing; Rothia.
© 2021 The Authors. Published by Elsevier Ltd.
Figures


References
-
- Ahluwalia M., Brailsford S.R., Tarelli E., Gilbert S.C., Clark D.T., Barnard K., Beighton D. Dental caries, oral hygiene, and oral clearance in children with craniofacial disorders. J. Dent. Res. 2004;83:175–179. - PubMed
-
- Bragger U., Schurch E., Jr., Gusberti F.A., Lang N.P. Periodontal conditions in adolescents with cleft lip, alveolus and palate following treatment in a co-ordinated team approach. J. Clin. Periodontol. 1985;12:494–502. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

