KRAS mutant rectal cancer cells interact with surrounding fibroblasts to deplete the extracellular matrix
- PMID: 33817986
- PMCID: PMC8486594
- DOI: 10.1002/1878-0261.12960
KRAS mutant rectal cancer cells interact with surrounding fibroblasts to deplete the extracellular matrix
Abstract
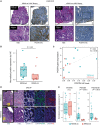
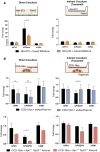
Somatic mutations in the KRAS oncogene are associated with poor outcomes in locally advanced rectal cancer but the underlying biologic mechanisms are not fully understood. We profiled mRNA in 76 locally advanced rectal adenocarcinomas from patients that were enrolled in a prospective clinical trial and investigated differences in gene expression between KRAS mutant (KRAS-mt) and KRAS-wild-type (KRAS-wt) patients. We found that KRAS-mt tumors display lower expression of genes related to the tumor stroma and remodeling of the extracellular matrix. We validated our findings using samples from The Cancer Genome Atlas (TCGA) and also by performing immunohistochemistry (IHC) and immunofluorescence (IF) in orthogonal cohorts. Using in vitro and in vivo models, we show that oncogenic KRAS signaling within the epithelial cancer cells modulates the activity of the surrounding fibroblasts in the tumor microenvironment.
Keywords: KRAS; cancer-associated fibroblast; extracellular matrix; rectal cancer; tumor response; tumor stroma.
© 2021 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
JGA has received honorarium for being a consultant with the following: Medtronics, Ethicon J&J, Da Vinci Intuitive Surgical. JJS has received travel support from Intuitive Surgical Inc. for fellow education (2015) and has served as a clinical advisor for Guardant Health, Inc (2019). The other co‐authors have no conflicts of interest to disclose.
Figures


References
-
- Siegel RL, Miller KD, Fuchs HE & Jemal A (2021) Cancer Statistics, 2021. CA Cancer J Clin 71, 7–33. - PubMed
-
- Andreyev HJ, Norman AR, Clarke PA, Cunningham D & Oates JR (1998) Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst 90, 675–684. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

