The Atg16l1 gene: characterization of wild type, knock-in, and knock-out phenotypes in rats
- PMID: 33818130
- PMCID: PMC8285577
- DOI: 10.1152/physiolgenomics.00114.2020
The Atg16l1 gene: characterization of wild type, knock-in, and knock-out phenotypes in rats
Abstract
ATG16L1 is a ubiquitous autophagy gene responsible, in part, for formation of the double-membrane bound autophagosome that delivers unwanted cellular debris and intracellular pathogens to the lysosome for degradation. A single, nonsynonymous adenine to guanine polymorphism resulting in a threonine to alanine amino acid substitution (T300A) directly preceded by a caspase cleavage site (DxxD) causes an increased susceptibility to Crohn's disease (CD) in humans. The mechanism behind this increased susceptibility is still being elucidated, however, the amino acid change caused by this point mutation results in increased ATG16L1 protein sensitivity to caspase 3-mediated cleavage. To generate novel rat strains carrying genetic alterations in the rat Atg16l1 gene, we first characterized the wild-type rat gene. We identified four alternative splice variants with tissue-specific expression. Using CRISPR-Cas9 genome editing technology, we developed a knock-in rat model for the human ATG16L1 T300A CD risk polymorphism, as well as a knock-out rat model to evaluate the role of Atg16l1 in autophagy as well as its potential effect on CD susceptibility. These are the first reported rat strains with alterations of the Atg16l1 gene. Consistent with studies of the effects of human ATG16L1 polymorphisms, models exhibit morphological abnormalities in both Paneth and goblet cells, but do not develop spontaneous intestinal permeability or inflammatory bowel disease. Analysis of the gut microbiota does not show inherent differences in bacterial composition between wild-type and genetically modified animals. These Atg16l1 strains are valuable new animal models for the study of both autophagy and CD susceptibility.
Keywords: CRISPR; Crohn's disease; genetic models; inflammatory bowel disease; phenotyping.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures



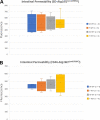


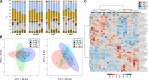
Similar articles
-
Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense.Proc Natl Acad Sci U S A. 2014 May 27;111(21):7741-6. doi: 10.1073/pnas.1407001111. Epub 2014 May 12. Proc Natl Acad Sci U S A. 2014. PMID: 24821797 Free PMC article.
-
A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3.Nature. 2014 Feb 27;506(7489):456-62. doi: 10.1038/nature13044. Epub 2014 Feb 19. Nature. 2014. PMID: 24553140
-
IBD-Associated Atg16L1T300A Polymorphism Regulates Commensal Microbiota of the Intestine.Front Immunol. 2022 Jan 27;12:772189. doi: 10.3389/fimmu.2021.772189. eCollection 2021. Front Immunol. 2022. PMID: 35154071 Free PMC article.
-
Classification of genetic profiles of Crohn's disease: a focus on the ATG16L1 gene.Expert Rev Mol Diagn. 2008 Mar;8(2):199-207. doi: 10.1586/14737159.8.2.199. Expert Rev Mol Diagn. 2008. PMID: 18366306 Review.
-
ATG16L1: A multifunctional susceptibility factor in Crohn disease.Autophagy. 2015 Apr 3;11(4):585-94. doi: 10.1080/15548627.2015.1017187. Autophagy. 2015. PMID: 25906181 Free PMC article. Review.
Cited by
-
Recent Advances in the Production of Genome-Edited Rats.Int J Mol Sci. 2022 Feb 25;23(5):2548. doi: 10.3390/ijms23052548. Int J Mol Sci. 2022. PMID: 35269691 Free PMC article. Review.
-
The relationship between extreme inter-individual variation in macrophage gene expression and genetic susceptibility to inflammatory bowel disease.Hum Genet. 2024 Mar;143(3):233-261. doi: 10.1007/s00439-024-02642-9. Epub 2024 Feb 29. Hum Genet. 2024. PMID: 38421405 Free PMC article.
-
Genetic and Epigenetic Etiology of Inflammatory Bowel Disease: An Update.Genes (Basel). 2022 Dec 16;13(12):2388. doi: 10.3390/genes13122388. Genes (Basel). 2022. PMID: 36553655 Free PMC article. Review.
-
Research progress on the relationship between Paneth cells-susceptibility genes, intestinal microecology and inflammatory bowel disease.World J Clin Cases. 2023 Dec 6;11(34):8111-8125. doi: 10.12998/wjcc.v11.i34.8111. World J Clin Cases. 2023. PMID: 38130785 Free PMC article. Review.
-
Main genetic factors associated with inflammatory bowel diseases and their consequences on intestinal permeability: involvement in gut inflammation.J Gastroenterol. 2025 Aug 21. doi: 10.1007/s00535-025-02289-x. Online ahead of print. J Gastroenterol. 2025. PMID: 40841786 Review.
References
-
- Cleynen I, González JR, Figueroa C, Franke A, McGovern D, Bortlík M, Crusius BJA, Vecchi M, Artieda M, Szczypiorska M, Bethge J, Arteta D, Ayala E, Danese S, van Hogezand RA, Panés J, Peña SA, Lukas M, Jewell DP, Schreiber S, Vermeire S, Sans M. Genetic factors conferring an increased susceptibility to develop Crohn’s disease also influence disease phenotype: results from the IBDchip European Project. Gut 62: 1556–1565, 2013. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

