Diverse genetic causes of polymicrogyria with epilepsy
- PMID: 33818783
- PMCID: PMC10838185
- DOI: 10.1111/epi.16854
Diverse genetic causes of polymicrogyria with epilepsy
Erratum in
-
Erratum.Epilepsia. 2021 Aug;62(8):2017. doi: 10.1111/epi.16960. Epub 2021 Jun 27. Epilepsia. 2021. PMID: 34180061 No abstract available.
Abstract
Objective: We sought to identify novel genes and to establish the contribution of known genes in a large cohort of patients with nonsyndromic sporadic polymicrogyria and epilepsy.
Methods: We enrolled participants with polymicrogyria and their parents through the Epilepsy Phenome/Genome Project. We performed phenotyping and whole exome sequencing (WES), trio analysis, and gene-level collapsing analysis to identify de novo or inherited variants, including germline or mosaic (postzygotic) single nucleotide variants, small insertion-deletion (indel) variants, and copy number variants present in leukocyte-derived DNA.
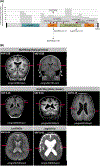
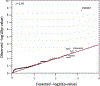
Results: Across the cohort of 86 individuals with polymicrogyria and epilepsy, we identified seven with pathogenic or likely pathogenic variants in PIK3R2, including four germline and three mosaic variants. PIK3R2 was the only gene harboring more than expected de novo variants across the entire cohort, and likewise the only gene that passed the genome-wide threshold of significance in the gene-level rare variant collapsing analysis. Consistent with previous reports, the PIK3R2 phenotype consisted of bilateral polymicrogyria concentrated in the perisylvian region with macrocephaly. Beyond PIK3R2, we also identified one case each with likely causal de novo variants in CCND2 and DYNC1H1 and biallelic variants in WDR62, all genes previously associated with polymicrogyria. Candidate genetic explanations in this cohort included single nucleotide de novo variants in other epilepsy-associated and neurodevelopmental disease-associated genes (SCN2A in two individuals, GRIA3, CACNA1C) and a 597-kb deletion at 15q25, a neurodevelopmental disease susceptibility locus.
Significance: This study confirms germline and postzygotically acquired de novo variants in PIK3R2 as an important cause of bilateral perisylvian polymicrogyria, notably with macrocephaly. In total, trio-based WES identified a genetic diagnosis in 12% and a candidate diagnosis in 6% of our polymicrogyria cohort. Our results suggest possible roles for SCN2A, GRIA3, CACNA1C, and 15q25 deletion in polymicrogyria, each already associated with epilepsy or other neurodevelopmental conditions without brain malformations. The role of these genes in polymicrogyria will be further understood as more patients with polymicrogyria undergo genetic evaluation.
Keywords: de novo variant; epilepsy; exome sequencing; polymicrogyria; trio.
© 2021 International League Against Epilepsy.
Conflict of interest statement
CONFLICT OF INTEREST
D.B.G. is a founder of and holds equity in Q State Biosciences and Praxis Therapeutics; holds equity in Apostle, and serves as a consultant to AstraZeneca and Gilead. S.Petrov. has equity in and is employed by AstraZeneca. O.D. receives grant support from NINDS, NIMH, MURI, CDC, and NSF. He has equity and/or compensation from the following companies: Privateer Holdings, Tilray, Receptor Life Sciences, Qstate Biosciences, Tevard, Empatica, Egg Rock/Papa & Barkley, Rettco, SilverSpike, and California Cannabis Enterprises. He has received consulting fees from GW Pharma, Cavion, Zogenix, and Eisai. R.A.S. receives research funding from PCORI, NIH, the Pediatric Epilepsy Research Foundation, and the University of Michigan. She serves as a consultant for the Epilepsy Study Consortium, receives royalties from UpToDate for authorship of topics related to neonatal seizures, and serves as an Associate Editor for
Figures

References
-
- Barkovich AJ, Kuzniecky RI, Jackson GD, Guerrini R, Dobyns WB. A developmental and genetic classification for malformations of cortical development. Neurology. 2005;65(12):1873–87. - PubMed
-
- Kammoun F, Tanguy A, Boesplug- Tanguy O, Bensahel H, Khouri N, Landrieu P. Club feet with congenital perisylvian polymicrogyria possibly due to bifocal ischemic damage of the neuraxis in utero. Am J Med Genet. 2004;126A(2):191–6. - PubMed
-
- Nissenkorn A, Michelson M, Ben- Zeev B, Lerman-Sagie T. Inborn errors of metabolism: a cause of abnormal brain development. Neurology. 2001;56(10):1265–7 2. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K01 MH098126/MH/NIMH NIH HHS/United States
- R56 AI098588/AI/NIAID NIH HHS/United States
- RC2 MH089915/MH/NIMH NIH HHS/United States
- R01 MH097971/MH/NIMH NIH HHS/United States
- R01 MH099216/MH/NIMH NIH HHS/United States
- R01 AG037212/AG/NIA NIH HHS/United States
- R01 DK080099/DK/NIDDK NIH HHS/United States
- UL1 TR000040/TR/NCATS NIH HHS/United States
- UL1 TR001873/TR/NCATS NIH HHS/United States
- R01 NS058721/NS/NINDS NIH HHS/United States
- MR/S02638X/1/MRC_/Medical Research Council/United Kingdom
- UM1 AI100645/AI/NIAID NIH HHS/United States
- P30 AG028377/AG/NIA NIH HHS/United States
- U01 NS077274/NS/NINDS NIH HHS/United States
- UM1 HG006504/HG/NHGRI NIH HHS/United States
- U54 NS078059/NS/NINDS NIH HHS/United States
- P01 AG007232/AG/NIA NIH HHS/United States
- RF1 AG054023/AG/NIA NIH HHS/United States
- R01 HD048805/HD/NICHD NIH HHS/United States
- U01 HG007672/HG/NHGRI NIH HHS/United States
- U01 NS053998/NS/NINDS NIH HHS/United States
- U01 NS077364/NS/NINDS NIH HHS/United States
- P01 HD080642/HD/NICHD NIH HHS/United States
- U01 NS077303/NS/NINDS NIH HHS/United States
- U19 AI067854/AI/NIAID NIH HHS/United States
- RC2 NS070344/NS/NINDS NIH HHS/United States
- U01 NS077276/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

