Bone Morphogenetic Protein Pathway Antagonism by Grem1 Regulates Epithelial Cell Fate in Intestinal Regeneration
- PMID: 33819486
- PMCID: PMC7613733
- DOI: 10.1053/j.gastro.2021.03.052
Bone Morphogenetic Protein Pathway Antagonism by Grem1 Regulates Epithelial Cell Fate in Intestinal Regeneration
Abstract
Background & aims: In homeostasis, intestinal cell fate is controlled by balanced gradients of morphogen signaling. The bone morphogenetic protein (BMP) pathway has a physiological, prodifferentiation role, predominantly inferred through previous experimental pathway inactivation. Intestinal regeneration is underpinned by dedifferentiation and cell plasticity, but the signaling pathways that regulate this adaptive reprogramming are not well understood. We assessed the BMP signaling landscape and investigated the impact and therapeutic potential of pathway manipulation in homeostasis and regeneration.
Methods: A novel mouse model was generated to assess the effect of the autocrine Bmp4 ligand on individual secretory cell fate. We spatiotemporally mapped BMP signaling in mouse and human regenerating intestine. Transgenic models were used to explore the functional impact of pathway manipulation on stem cell fate and intestinal regeneration.
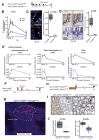
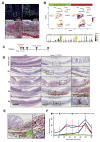
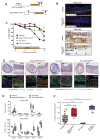
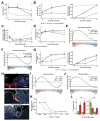
Results: In homeostasis, ligand exposure reduced proliferation, expedited terminal differentiation, abrogated secretory cell survival, and prevented dedifferentiation. After ulceration, physiological attenuation of BMP signaling arose through upregulation of the secreted antagonist Grem1 from topographically distinct populations of fibroblasts. Concomitant expression supported functional compensation after Grem1 deletion from tissue-resident cells. BMP pathway manipulation showed that antagonist-mediated BMP attenuation was obligatory but functionally submaximal, because regeneration was impaired or enhanced by epithelial overexpression of Bmp4 or Grem1, respectively. Mechanistically, Bmp4 abrogated regenerative stem cell reprogramming despite a convergent impact of YAP/TAZ on cell fate in remodeled wounds.
Conclusions: BMP signaling prevents epithelial dedifferentiation, and pathway attenuation through stromal Grem1 upregulation was required for adaptive reprogramming in intestinal regeneration. This intercompartmental antagonism was functionally submaximal, raising the possibility of therapeutic pathway manipulation in inflammatory bowel disease.
Keywords: Bone Morphogenetic Protein; Dedifferentiation; Grem1; Intestinal Regeneration.
Copyright © 2021 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
These authors disclose the following: S.J.L. has received grant income from UCB Pharma. V.H.K. has served as an invited speaker on behalf of Indica Labs. The remaining authors disclose no conflicts.
Figures


References
-
- Scoville D, Sato T, He X, et al. Current view: intestinal stem cells and signaling. Gastroenterology. 2008;134:849–864. - PubMed
-
- van den Brink G, Offerhaus G. The morphogenetic code and colon cancer development. Cancer Cell. 2007;11:109–117. - PubMed
-
- He XC, Zhang J, Tong WG, et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat Genet. 2004;36:1117–1121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

