The Small Molecule H89 Inhibits Chlamydia Inclusion Growth and Production of Infectious Progeny
- PMID: 33820812
- PMCID: PMC8373235
- DOI: 10.1128/IAI.00729-20
The Small Molecule H89 Inhibits Chlamydia Inclusion Growth and Production of Infectious Progeny
Abstract
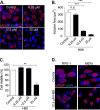
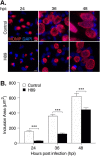
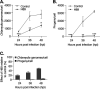
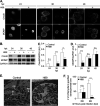
Chlamydia is an obligate intracellular bacterium and the most common reportable cause of human infection in the United States. This pathogen proliferates inside a eukaryotic host cell, where it resides within a membrane-bound compartment called the chlamydial inclusion. It has an unusual developmental cycle, marked by conversion between a replicating form, the reticulate body (RB), and an infectious form, the elementary body (EB). We found that the small molecule H89 slowed inclusion growth and decreased overall RB replication by 2-fold but caused a 25-fold reduction in infectious EBs. This disproportionate effect on EB production was mainly due to a defect in RB-to-EB conversion and not to the induction of chlamydial persistence, which is an altered growth state. Although H89 is a known inhibitor of specific protein kinases and vesicular transport to and from the Golgi apparatus, it did not cause these anti-chlamydial effects by blocking protein kinase A or C or by inhibiting protein or lipid transport. Thus, H89 is a novel anti-chlamydial compound that has a unique combination of effects on an intracellular Chlamydia infection.
Keywords: RB-to-EB conversion; developmental cycle; intracellular infection; isoquinoline sulfonamide.
Figures

Similar articles
-
Differential Effects of Small Molecule Inhibitors on the Intracellular Chlamydia Infection.mBio. 2022 Aug 30;13(4):e0107622. doi: 10.1128/mbio.01076-22. Epub 2022 Jun 15. mBio. 2022. PMID: 35703434 Free PMC article.
-
The chlamydial transcriptional regulator Euo is a key switch in cell form developmental progression but is not involved in the committed step to the formation of the infectious form.mSphere. 2024 Sep 25;9(9):e0043724. doi: 10.1128/msphere.00437-24. Epub 2024 Aug 14. mSphere. 2024. PMID: 39140730 Free PMC article.
-
The Periplasmic Tail-Specific Protease, Tsp, Is Essential for Secondary Differentiation in Chlamydia trachomatis.J Bacteriol. 2023 May 25;205(5):e0009923. doi: 10.1128/jb.00099-23. Epub 2023 Apr 24. J Bacteriol. 2023. PMID: 37092988 Free PMC article.
-
Targeting eukaryotic Rab proteins: a smart strategy for chlamydial survival and replication.Cell Microbiol. 2014 Sep;16(9):1329-38. doi: 10.1111/cmi.12325. Epub 2014 Aug 4. Cell Microbiol. 2014. PMID: 24948448 Review.
-
Type III Secretion in Chlamydia.Microbiol Mol Biol Rev. 2023 Sep 26;87(3):e0003423. doi: 10.1128/mmbr.00034-23. Epub 2023 Jun 26. Microbiol Mol Biol Rev. 2023. PMID: 37358451 Free PMC article. Review.
Cited by
-
Development of an sRNA-mediated conditional knockdown system for Chlamydia trachomatis.mBio. 2025 Feb 5;16(2):e0254524. doi: 10.1128/mbio.02545-24. Epub 2024 Dec 13. mBio. 2025. PMID: 39670716 Free PMC article.
-
Differential Effects of Small Molecule Inhibitors on the Intracellular Chlamydia Infection.mBio. 2022 Aug 30;13(4):e0107622. doi: 10.1128/mbio.01076-22. Epub 2022 Jun 15. mBio. 2022. PMID: 35703434 Free PMC article.
-
Chlamydia trachomatis induces disassembly of the primary cilium to promote the intracellular infection.PLoS Pathog. 2024 Jun 17;20(6):e1012303. doi: 10.1371/journal.ppat.1012303. eCollection 2024 Jun. PLoS Pathog. 2024. PMID: 38885287 Free PMC article.
-
Alternative strategies for Chlamydia treatment: Promising non-antibiotic approaches.Front Microbiol. 2022 Nov 23;13:987662. doi: 10.3389/fmicb.2022.987662. eCollection 2022. Front Microbiol. 2022. PMID: 36504792 Free PMC article. Review.
-
A Reverse Genetic Approach for Studying sRNAs in Chlamydia trachomatis.mBio. 2022 Aug 30;13(4):e0086422. doi: 10.1128/mbio.00864-22. Epub 2022 Jun 21. mBio. 2022. PMID: 35726915 Free PMC article.
References
-
- Centers for Disease Control and Prevention. 2019. CDC–Chlamydia statistics. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/std/chlamydia/stats.htm.
-
- Batteiger BE, Tan M. 2019. Chlamydia trachomatis (trachoma and urogenital infections), p 2301–2319. In Principles and practice of infectious diseases. Elsevier Inc., Philadelphia, PA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

