Genome-Wide Analysis of Experimentally Evolved Candida auris Reveals Multiple Novel Mechanisms of Multidrug Resistance
- PMID: 33820824
- PMCID: PMC8092288
- DOI: 10.1128/mBio.03333-20
Genome-Wide Analysis of Experimentally Evolved Candida auris Reveals Multiple Novel Mechanisms of Multidrug Resistance
Abstract
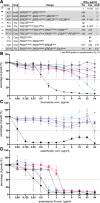
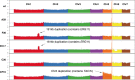
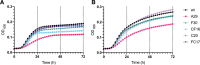
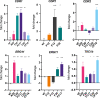
Candida auris is globally recognized as an opportunistic fungal pathogen of high concern, due to its extensive multidrug resistance (MDR). Still, molecular mechanisms of MDR are largely unexplored. This is the first account of genome-wide evolution of MDR in C. auris obtained through serial in vitro exposure to azoles, polyenes, and echinocandins. We show the stepwise accumulation of copy number variations and novel mutations in genes both known and unknown in antifungal drug resistance. Echinocandin resistance was accompanied by a codon deletion in FKS1 hot spot 1 and a substitution in FKS1 "novel" hot spot 3. Mutations in ERG3 and CIS2 further increased the echinocandin MIC. Decreased azole susceptibility was linked to a mutation in transcription factor TAC1b and overexpression of the drug efflux pump Cdr1, a segmental duplication of chromosome 1 containing ERG11, and a whole chromosome 5 duplication, which contains TAC1b The latter was associated with increased expression of ERG11, TAC1b, and CDR2 but not CDR1 The simultaneous emergence of nonsense mutations in ERG3 and ERG11 was shown to decrease amphotericin B susceptibility, accompanied with fluconazole cross-resistance. A mutation in MEC3, a gene mainly known for its role in DNA damage homeostasis, further increased the polyene MIC. Overall, this study shows the alarming potential for and diversity of MDR development in C. auris, even in a clade until now not associated with MDR (clade II), stressing its clinical importance and the urge for future research.IMPORTANCECandida auris is a recently discovered human fungal pathogen and has shown an alarming potential for developing multi- and pan-resistance toward all classes of antifungals most commonly used in the clinic. Currently, C. auris has been globally recognized as a nosocomial pathogen of high concern due to this evolutionary potential. So far, this is the first study in which the stepwise progression of multidrug resistance (MDR) in C. auris is monitored in vitro Multiple novel mutations in known resistance genes and genes previously not or vaguely associated with drug resistance reveal rapid MDR evolution in a C. auris clade II isolate. Additionally, this study shows that in vitro experimental evolution can be a powerful tool to discover new drug resistance mechanisms, although it has its limitations.
Keywords: Candida auris; amphotericin B; antifungal agents; caspofungin; drug resistance evolution; experimental evolution; fluconazole; genome analysis; microevolution; multidrug resistance; whole-genome sequencing.
Copyright © 2021 Carolus et al.
Figures


References
-
- CDC. 2019. Antibiotic Resistance Threats in the United States, 2019. www.cdc.gov/DrugResistance/Biggest-Threats.html. Accessed 26 March 2020.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

