A method for detection of SARS-CoV-2 RNA in healthy human stool: a validation study
- PMID: 33821247
- PMCID: PMC8012028
- DOI: 10.1016/S2666-5247(21)00059-8
A method for detection of SARS-CoV-2 RNA in healthy human stool: a validation study
Abstract
Background: Faecal shedding of SARS-CoV-2 has raised concerns about transmission through faecal microbiota transplantation procedures. Validation parameters of authorised tests for SARS-CoV-2 RNA detection in respiratory samples are described in product labelling, whereas the published methods for SARS-CoV-2 detection from faecal samples have not permitted a robust description of the assay parameters. We aimed to develop and validate a test specifically for detection of SARS-CoV-2 in human stool.
Methods: In this validation study, we evaluated performance characteristics of a reverse transcriptase real-time PCR (RT-rtPCR) test for detection of SARS-CoV-2 in human stool specimens by spiking stool with inactivated SARS-CoV-2 material. A modified version of the US Centers for Disease Control and Prevention RT-rtPCR SARS-CoV-2 test was used for detection of viral RNA. Analytical sensitivity was evaluated in freshly spiked stool by testing two-fold dilutions in replicates of 20. Masked samples were tested by a second laboratory to evaluate interlaboratory reproducibility. Short-term (7-day) stability of viral RNA in stool samples was assessed with four different stool storage buffers (phosphate-buffered saline, Cary-Blair medium, Stool Transport and Recovery [STAR] buffer, and DNA/RNA Shield) kept at -80°C, 4°C, and ambient temperature (approximately 21°C). We also tested clinical stool and anal swab specimens from patients who were SARS-CoV-2 positive by nasopharyngeal testing.
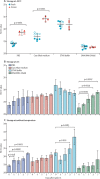
Findings: The lower limit of detection of the assay was found to be 3000 viral RNA copies per g of original stool sample, with 100% detection across 20 replicates assessed at this concentration. Analytical sensitivity was diminished by approximately two times after a single freeze-thaw cycle at -80°C. At 100 times the limit of detection, spiked samples were generally stable in all four stool storage buffers tested for up to 7 days, with maximum changes in mean threshold cycle values observed at -80°C storage in Cary-Blair medium (from 29·4 [SD 0·27] at baseline to 30·8 [0·17] at day 7; p<0·0001), at 4°C storage in DNA/RNA Shield (from 28·5 [0·15] to 29·8 [0·09]; p=0·0019), and at ambient temperature in STAR buffer (from 30·4 [0·24] to 32·4 [0·62]; p=0·0083). 30 contrived SARS-CoV-2 samples were tested by a second laboratory and were correctly identified as positive or negative in at least one of two rounds of testing. Additionally, SARS-CoV-2 RNA was detected using this assay in the stool and anal swab specimens of 11 of 23 individuals known to be positive for SARS-CoV-2.
Interpretation: This is a sensitive and reproducible assay for detection of SARS-CoV-2 RNA in human stool, with potential uses in faecal microbiota transplantation donor screening, sewage monitoring, and further research into the effects of faecal shedding on the epidemiology of the COVID-19 pandemic.
Funding: National Institute of Allergy and Infectious Diseases, US National Institutes of Health; Center for Biologics Evaluation and Research, US Food and Drug Administration.
© 2021 Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license.
Conflict of interest statement
PEC reports grants from the National Institute of Allergy and Infectious Diseases, US National Institutes of Health, during the conduct of the study. All other authors declare no competing interests.
Figures
References
-
- Carlson PE., Jr Regulatory considerations for fecal microbiota transplantation products. Cell Host Microbe. 2020;27:173–175. - PubMed
-
- US Food and Drug Administration Information pertaining to additional safety protections regarding use of fecal microbiota for transplantation—testing of stool donors for enteropathogenic Escherichia coli and Shigatoxin-producing Escherichia coli. April 6, 2020. https://www.fda.gov/vaccines-blood-biologics/safety-availability-biologi...
-
- US Food and Drug Administration Safety alert regarding use of fecal microbiota for transplantation and risk of serious adverse events likely due to transmission of pathogenic organisms. March 12, 2020. https://www.fda.gov/vaccines-blood-biologics/safety-availability-biologi...
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

