A potent neutralizing nanobody against SARS-CoV-2 with inhaled delivery potential
- PMID: 33821254
- PMCID: PMC8013425
- DOI: 10.1002/mco2.60
A potent neutralizing nanobody against SARS-CoV-2 with inhaled delivery potential
Abstract
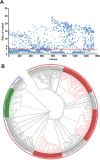
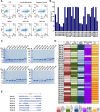
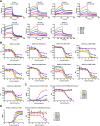
The coronavirus disease 2019 (COVID-19) pandemic has become a serious burden on global public health. Although therapeutic drugs against COVID-19 have been used in many countries, their efficacy is still limited. We here reported nanobody (Nb) phage display libraries derived from four camels immunized with the SARS-CoV-2 spike receptor-binding domain (RBD), from which 381 Nbs were identified to recognize SARS-CoV-2-RBD. Furthermore, seven Nbs were shown to block interaction of human angiotensin-converting enzyme 2 (ACE2) with SARS-CoV-2-RBD variants and two Nbs blocked the interaction of human ACE2 with bat-SL-CoV-WIV1-RBD and SARS-CoV-1-RBD. Among these candidates, Nb11-59 exhibited the highest activity against authentic SARS-CoV-2 with 50% neutralizing dose (ND50) of 0.55 μg/ml. Nb11-59 can be produced on large scale in Pichia pastoris, with 20 g/L titer and 99.36% purity. It also showed good stability profile, and nebulization did not impact its stability. Overall, Nb11-59 might be a promising prophylactic and therapeutic molecule against COVID-19, especially through inhalation delivery.
Keywords: SARS‐CoV‐2; large‐scale production; nanobody; nebulization; neutralizing activity.
© 2021 The Authors. MedComm published by Sichuan International Medical Exchange & Promotion Association (SCIMEA) and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
All commercial rights from this paper belong to Shanghai Novamab Biopharmaceuticals Co., Ltd.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
