This is a preprint.
Severity of SARS-CoV-2 infection as a function of the interferon landscape across the respiratory tract of COVID-19 patients
- PMID: 33821280
- PMCID: PMC8020981
- DOI: 10.1101/2021.03.30.437173
Severity of SARS-CoV-2 infection as a function of the interferon landscape across the respiratory tract of COVID-19 patients
Update in
-
The interferon landscape along the respiratory tract impacts the severity of COVID-19.Cell. 2021 Sep 16;184(19):4953-4968.e16. doi: 10.1016/j.cell.2021.08.016. Epub 2021 Aug 19. Cell. 2021. PMID: 34492226 Free PMC article.
Abstract
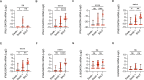
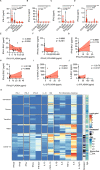
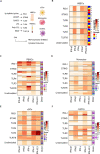
The COVID-19 outbreak driven by SARS-CoV-2 has caused more than 2.5 million deaths globally, with the most severe cases characterized by over-exuberant production of immune-mediators, the nature of which is not fully understood. Interferons of the type I (IFN-I) or type III (IFN-III) families are potent antivirals, but their role in COVID-19 remains debated. Our analysis of gene and protein expression along the respiratory tract shows that IFNs, especially IFN-III, are over-represented in the lower airways of patients with severe COVID-19, while high levels of IFN-III, and to a lesser extent IFN-I, characterize the upper airways of patients with high viral burden but reduced disease risk or severity; also, IFN expression varies with abundance of the cell types that produce them. Our data point to a dynamic process of inter- and intra-family production of IFNs in COVID-19, and suggest that IFNs play opposing roles at distinct anatomical sites.
Conflict of interest statement
Declaration of Interests.
IZ reports compensation for consulting services with Implicit Biosciences.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
