A Cytotoxic Bis(1,2,3-triazol-5-ylidene)carbazolide Gold(III) Complex Targets DNA by Partial Intercalation
- PMID: 33822431
- PMCID: PMC8251726
- DOI: 10.1002/chem.202100598
A Cytotoxic Bis(1,2,3-triazol-5-ylidene)carbazolide Gold(III) Complex Targets DNA by Partial Intercalation
Abstract
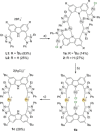
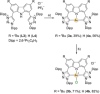
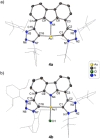
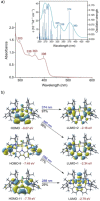
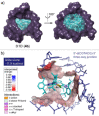
The syntheses of bis(triazolium)carbazole precursors and their corresponding coinage metal (Au, Ag) complexes are reported. For alkylated triazolium salts, di- or tetranuclear complexes with bridging ligands were isolated, while the bis(aryl) analogue afforded a bis(carbene) AuI -CNC pincer complex suitable for oxidation to the redox-stable [AuIII (CNC)Cl]+ cation. Although the ligand salt and the [AuIII (CNC)Cl]+ complex were both notably cytotoxic toward the breast cancer cell line MDA-MB-231, the AuIII complex was somewhat more selective. Electrophoresis, viscometry, UV-vis, CD and LD spectroscopy suggest the cytotoxic [AuIII (CNC)Cl]+ complex behaves as a partial DNA intercalator. In silico screening indicated that the [AuIII (CNC)Cl]+ complex can target DNA three-way junctions with good specificity, several other regular B-DNA forms, and Z-DNA. Multiple hydrophobic π-type interactions involving T and A bases appear to be important for B-form DNA binding, while phosphate O⋅⋅⋅Au interactions evidently underpin Z-DNA binding. The CNC ligand effectively stabilizes the AuIII ion, preventing reduction in the presence of glutathione. Both the redox stability and DNA affinity of the hit compound might be key factors underpinning its cytotoxicity in vitro.
Keywords: anticancer; cytotoxicity; gold complexes; mesoionic carbenes; metallodrugs.
© 2021 The Authors. Chemistry - A European Journal published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Simpson P. V., Desai N. M., Casari I., Massi M., Falasca M., Future Med. Chem. 2019, 11, 119–135. - PubMed
-
- Allardyce C. S., Dyson P. J., Dalton Trans. 2016, 45, 3201–3209. - PubMed
-
- Gianferrara T., Bratsos I., Alessio E., Dalton Trans. 2009, 7588–7598. - PubMed
-
- Fricker S. P., Dalton Trans. 2007, 4903–4917. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

