JMJD6 Is a Druggable Oxygenase That Regulates AR-V7 Expression in Prostate Cancer
- PMID: 33822745
- PMCID: PMC8025710
- DOI: 10.1158/0008-5472.CAN-20-1807
JMJD6 Is a Druggable Oxygenase That Regulates AR-V7 Expression in Prostate Cancer
Abstract
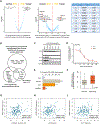
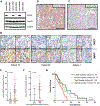
Endocrine resistance (EnR) in advanced prostate cancer is fatal. EnR can be mediated by androgen receptor (AR) splice variants, with AR splice variant 7 (AR-V7) arguably the most clinically important variant. In this study, we determined proteins key to generating AR-V7, validated our findings using clinical samples, and studied splicing regulatory mechanisms in prostate cancer models. Triangulation studies identified JMJD6 as a key regulator of AR-V7, as evidenced by its upregulation with in vitro EnR, its downregulation alongside AR-V7 by bromodomain inhibition, and its identification as a top hit of a targeted siRNA screen of spliceosome-related genes. JMJD6 protein levels increased (P < 0.001) with castration resistance and were associated with higher AR-V7 levels and shorter survival (P = 0.048). JMJD6 knockdown reduced prostate cancer cell growth, AR-V7 levels, and recruitment of U2AF65 to AR pre-mRNA. Mutagenesis studies suggested that JMJD6 activity is key to the generation of AR-V7, with the catalytic machinery residing within a druggable pocket. Taken together, these data highlight the relationship between JMJD6 and AR-V7 in advanced prostate cancer and support further evaluation of JMJD6 as a therapeutic target in this disease. SIGNIFICANCE: This study identifies JMJD6 as being critical for the generation of AR-V7 in prostate cancer, where it may serve as a tractable target for therapeutic intervention.
©2021 American Association for Cancer Research.
Conflict of interest statement
Conflicts of interest
AP, JW, AN, WY, IF, RP, AF, RR, DNR, PDM, BG, RC, VG, LB, MC, SM, MBL, SC, NT, BAL, AS and JdB are all employees of The Institute of Cancer Research (ICR), which has a commercial interest in abiraterone. The ICR operates a Rewards to Inventors scheme through which employees of the ICR may receive financial benefit following commercial licensing. BAL is currently or has been a consultant and received fees from to Astex Pharmaceuticals, GSK and Difiniens AG (member of Astra Zeneca group) and received speakers honorarium from Astellas Pharma. BAL is a former employee of Inpharmatica Ltd. SRP is President of ESSA Pharma (Consultant), ProsTech Inc. AS has received travel support from Sanofi and Roche-Genentech, and speakers honorarium from Astellas Pharma. JdB has served on advisory boards and received fees from many companies including Astra Zeneca, Astellas, Bayer, Boehringer Ingelheim, Cellcentric, Daiichi, Genentech/Roche, Genmab, GSK, Janssen, Merck Serono, Merck Sharp & Dohme, Menarini/Silicon Biosystems, Orion, Pfizer, Qiagen, Sanofi Aventis, Sierra Oncology, Taiho, Vertex Pharmaceuticals. He is an employee of The ICR, which have received funding or other support for his research work from AZ, Astellas, Bayer, Cellcentric, Daiichi, Genentech, Genmab, GSK, Janssen, Merck Serono, MSD, Menarini/Silicon Biosystems, Orion, Sanofi Aventis, Sierra Oncology, Taiho, Pfizer, Vertex, and which has a commercial interest in abiraterone, PARP inhibition in DNA repair defective cancers and PI3K/AKT pathway inhibitors (no personal income). JdB was named as an inventor, with no financial interest, for patent 8,822,438. He has been the CI/PI of many industry sponsored clinical trials.
Figures



References
-
- Visakorpi T, et al., In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet, 1995. 9(4): p. 401–6. - PubMed
-
- Scher HI, et al., Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med, 2012. 367(13): p. 1187–97. - PubMed
-
- Fizazi K, et al., Abiraterone plus Prednisone in Metastatic, Castration-Sensitive Prostate Cancer. N Engl J Med, 2017. 377(4): p. 352–360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

