Ecological and Evolutionary responses to Antibiotic Treatment in the Human Gut Microbiota
- PMID: 33822937
- PMCID: PMC8498795
- DOI: 10.1093/femsre/fuab018
Ecological and Evolutionary responses to Antibiotic Treatment in the Human Gut Microbiota
Abstract
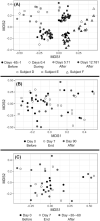
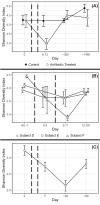
The potential for antibiotics to affect the ecology and evolution of the human gut microbiota is well recognised and has wide-ranging implications for host health. Here, we review the findings of key studies that surveyed the human gut microbiota during antibiotic treatment. We find several broad patterns including the loss of diversity, disturbance of community composition, suppression of bacteria in the Actinobacteria phylum, amplification of bacteria in the Bacteroidetes phylum, and promotion of antibiotic resistance. Such changes to the microbiota were often, but not always, recovered following the end of treatment. However, many studies reported unique and/or contradictory results, which highlights our inability to meaningfully predict or explain the effects of antibiotic treatment on the human gut microbiome. This problem arises from variation between existing studies in three major categories: differences in dose, class and combinations of antibiotic treatments used; differences in demographics, lifestyles, and locations of subjects; and differences in measurements, analyses and reporting styles used by researchers. To overcome this, we suggest two integrated approaches: (i) a top-down approach focused on building predictive models through large sample sizes, deep metagenomic sequencing, and effective collaboration; and (ii) a bottom-up reductionist approach focused on testing hypotheses using model systems.
Keywords: antibiotics; diversity; ecology; evolution; gut microbiota.
© The Author(s) 2021. Published by Oxford University Press on behalf of FEMS.
Figures


References
-
- Aminov R. History of antimicrobial drug discovery: major classes and health impact. Biochem Pharmacol. 2017;133:4–19. - PubMed
-
- Amábile-Cuevas CF, Chicurel ME. Bacterial plasmids and gene flux. Cell. 1992;70:189–99. - PubMed
-
- Andersson DI, Hughes D.. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol. 2014;12:465–78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

