Vaccination with prefusion-stabilized respiratory syncytial virus fusion protein induces genetically and antigenically diverse antibody responses
- PMID: 33823129
- PMCID: PMC8099422
- DOI: 10.1016/j.immuni.2021.03.004
Vaccination with prefusion-stabilized respiratory syncytial virus fusion protein induces genetically and antigenically diverse antibody responses
Abstract
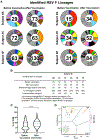
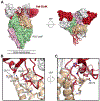
An effective vaccine for respiratory syncytial virus (RSV) is an unrealized public health goal. A single dose of the prefusion-stabilized fusion (F) glycoprotein subunit vaccine (DS-Cav1) substantially increases serum-neutralizing activity in healthy adults. We sought to determine whether DS-Cav1 vaccination induces a repertoire mirroring the pre-existing diversity from natural infection or whether antibody lineages targeting specific epitopes predominate. We evaluated RSV F-specific B cell responses before and after vaccination in six participants using complementary B cell sequencing methodologies and identified 555 clonal lineages. DS-Cav1-induced lineages recognized the prefusion conformation of F (pre-F) and were genetically diverse. Expressed antibodies recognized all six antigenic sites on the pre-F trimer. We identified 34 public clonotypes, and structural analysis of two antibodies from a predominant clonotype revealed a common mode of recognition. Thus, vaccination with DS-Cav1 generates a diverse polyclonal response targeting the antigenic sites on pre-F, supporting the development and advanced testing of pre-F-based vaccines against RSV.
Keywords: RSV; antibody repertoire; cryo-EM structure; fusion glycoprotein; memory B cells; neutralizing antibodies; prefusion; public clonotypes; respiratory syncytial virus.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests B.S.G., J.S.M., and P.D.K. are inventors on a patent entitled “Prefusion RSV F proteins and their use” (US Patent No. 9738689).
Figures



Similar articles
-
Alternative Virus-Like Particle-Associated Prefusion F Proteins as Maternal Vaccines for Respiratory Syncytial Virus.J Virol. 2019 Nov 13;93(23):e00914-19. doi: 10.1128/JVI.00914-19. Print 2019 Dec 1. J Virol. 2019. PMID: 31511382 Free PMC article.
-
Vaccination with prefusion-stabilized respiratory syncytial virus fusion protein elicits antibodies targeting a membrane-proximal epitope.J Virol. 2023 Oct 31;97(10):e0092923. doi: 10.1128/jvi.00929-23. Epub 2023 Sep 22. J Virol. 2023. PMID: 37737588 Free PMC article.
-
Respiratory syncytial virus prefusogenic fusion (F) protein nanoparticle vaccine: Structure, antigenic profile, immunogenicity, and protection.Vaccine. 2019 Sep 24;37(41):6112-6124. doi: 10.1016/j.vaccine.2019.07.089. Epub 2019 Aug 12. Vaccine. 2019. PMID: 31416644
-
Human respiratory syncytial virus: pathogenesis, immune responses, and current vaccine approaches.Eur J Clin Microbiol Infect Dis. 2018 Oct;37(10):1817-1827. doi: 10.1007/s10096-018-3289-4. Epub 2018 Jun 6. Eur J Clin Microbiol Infect Dis. 2018. PMID: 29876771 Review.
-
Clinical Potential of Prefusion RSV F-specific Antibodies.Trends Microbiol. 2018 Mar;26(3):209-219. doi: 10.1016/j.tim.2017.09.009. Epub 2017 Oct 17. Trends Microbiol. 2018. PMID: 29054341 Review.
Cited by
-
A single amino acid mutation alters multiple neutralization epitopes in the respiratory syncytial virus fusion glycoprotein.Npj Viruses. 2025 Apr 22;3(1):33. doi: 10.1038/s44298-025-00119-8. Npj Viruses. 2025. PMID: 40295799 Free PMC article.
-
Elicitation of pneumovirus-specific B cell responses by a prefusion-stabilized respiratory syncytial virus F subunit vaccine.Sci Transl Med. 2022 Jun 22;14(650):eabo5032. doi: 10.1126/scitranslmed.abo5032. Epub 2022 Jun 22. Sci Transl Med. 2022. PMID: 35731888 Free PMC article.
-
Construction and protective efficacy of a novel Streptococcus pneumoniae fusion protein vaccine NanAT1-TufT1-PlyD4.Front Immunol. 2022 Oct 31;13:1043293. doi: 10.3389/fimmu.2022.1043293. eCollection 2022. Front Immunol. 2022. PMID: 36389808 Free PMC article.
-
Broad anti-SARS-CoV-2 antibody immunity induced by heterologous ChAdOx1/mRNA-1273 vaccination.Science. 2022 Mar 4;375(6584):1041-1047. doi: 10.1126/science.abn2688. Epub 2022 Feb 10. Science. 2022. PMID: 35143256 Free PMC article.
-
Frequency-potency analysis of IgG+ memory B cells delineates neutralizing antibody responses at single-cell resolution.Cell Rep. 2024 Mar 26;43(3):113948. doi: 10.1016/j.celrep.2024.113948. Epub 2024 Mar 13. Cell Rep. 2024. PMID: 38483908 Free PMC article.
References
-
- Abbott RK, Lee JH, Menis S, Skog P, Rossi M, Ota T, Kulp DW, Bhullar D, Kalyuzhniy O, Havenar-Daughton C, et al. (2018). Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity 48, 133–146.e6. - PMC - PubMed
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, and Terwilliger TC (2002). PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr 58, 1948–1954. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

