Small molecule targeting of biologically relevant RNA tertiary and quaternary structures
- PMID: 33823146
- PMCID: PMC8141026
- DOI: 10.1016/j.chembiol.2021.03.003
Small molecule targeting of biologically relevant RNA tertiary and quaternary structures
Abstract
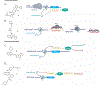
Initial successes in developing small molecule ligands for non-coding RNAs have underscored their potential as therapeutic targets. More recently, these successes have been aided by advances in biophysical and structural techniques for identification and characterization of more complex RNA structures; these higher-level folds present protein-like binding pockets that offer opportunities to design small molecules that could achieve a degree of selectivity often hard to obtain at the primary and secondary structure level. More specifically, identification and small molecule targeting of RNA tertiary and quaternary structures have allowed researchers to probe several human diseases and have resulted in promising clinical candidates. In this review we highlight a selection of diverse and exciting successes and the experimental approaches that led to their discovery. These studies include examples of recent developments in RNA-centric assays and ligands that provide insight into the features responsible for the affinity and biological outcome of RNA-targeted chemical probes. This report highlights the potential and emerging opportunities to selectively target RNA tertiary and quaternary structures as a route to better understand and, ultimately, treat many diseases.
Keywords: G-quadruplex; RNA structure; high-throughput screening; pseudoknot; quaternary; riboswitch; small molecule; synthetic library; tertiary; triple helix.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no conflict of interest.
Figures



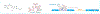
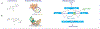

Similar articles
-
Small molecule-RNA targeting: starting with the fundamentals.Chem Commun (Camb). 2020 Nov 26;56(94):14744-14756. doi: 10.1039/d0cc06796b. Chem Commun (Camb). 2020. PMID: 33201954 Free PMC article.
-
Modulating RNA secondary and tertiary structures by mismatch binding ligands.Methods. 2019 Sep 1;167:78-91. doi: 10.1016/j.ymeth.2019.05.006. Epub 2019 May 10. Methods. 2019. PMID: 31078794 Review.
-
Identifying and validating small molecules interacting with RNA (SMIRNAs).Methods Enzymol. 2019;623:45-66. doi: 10.1016/bs.mie.2019.04.027. Epub 2019 May 15. Methods Enzymol. 2019. PMID: 31239057 Free PMC article.
-
Using SHAPE-MaP to probe small molecule-RNA interactions.Methods. 2019 Sep 1;167:105-116. doi: 10.1016/j.ymeth.2019.04.009. Epub 2019 Apr 19. Methods. 2019. PMID: 31009771 Review.
-
Fluorescence-based investigations of RNA-small molecule interactions.Methods. 2019 Sep 1;167:54-65. doi: 10.1016/j.ymeth.2019.05.017. Epub 2019 May 23. Methods. 2019. PMID: 31129289 Free PMC article.
Cited by
-
Thermal titration molecular dynamics (TTMD): shedding light on the stability of RNA-small molecule complexes.Front Mol Biosci. 2023 Nov 13;10:1294543. doi: 10.3389/fmolb.2023.1294543. eCollection 2023. Front Mol Biosci. 2023. PMID: 38028536 Free PMC article.
-
Using the structural diversity of RNA: protein interfaces to selectively target RNA with small molecules in cells: methods and perspectives.Front Mol Biosci. 2023 Nov 16;10:1298441. doi: 10.3389/fmolb.2023.1298441. eCollection 2023. Front Mol Biosci. 2023. PMID: 38033386 Free PMC article. Review.
-
Multiassay Profiling of a Focused Small Molecule Library Reveals Predictive Bidirectional Modulation of the lncRNA MALAT1 Triplex Stability In Vitro.ACS Chem Biol. 2022 Sep 16;17(9):2437-2447. doi: 10.1021/acschembio.2c00124. Epub 2022 Aug 19. ACS Chem Biol. 2022. PMID: 35984959 Free PMC article.
-
Computational drug discovery under RNA times.QRB Discov. 2022 Nov 14;3:e22. doi: 10.1017/qrd.2022.20. eCollection 2022. QRB Discov. 2022. PMID: 37529286 Free PMC article. Review.
-
Biomolecules Interacting with Long Noncoding RNAs.Biology (Basel). 2025 Apr 19;14(4):442. doi: 10.3390/biology14040442. Biology (Basel). 2025. PMID: 40282307 Free PMC article. Review.
References
-
- Arora A, and Maiti S (2009). Differential Biophysical Behavior of Human Telomeric RNA and DNA Quadruplex. The Journal of Physical Chemistry B 113, 10515–10520. - PubMed
-
- Bao H-L, and Xu Y (2018). Investigation of higher-order RNA G-quadruplex structures in vitro and in living cells by 19F NMR spectroscopy. Nature Protocols 13, 652–665. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

