Light-regulated pre-mRNA splicing in plants
- PMID: 33823333
- PMCID: PMC8487434
- DOI: 10.1016/j.pbi.2021.102037
Light-regulated pre-mRNA splicing in plants
Abstract
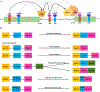
Light signal perceived by the red/far-red absorbing phytochrome (phy) family of photoreceptors regulates plant growth and development throughout the life cycle. Phytochromes regulate the light-triggered physiological responses by controlling gene expression both at the transcriptional and post-transcriptional levels. Recent large-scale RNA-seq studies have demonstrated the roles of phys in altering the global transcript diversity by modulating the pre-mRNA splicing in response to light. Moreover, several phy-interacting splicing factors/regulators from different species have been identified using forward genetics and protein-protein interaction studies, which modulate the light-regulated pre-mRNA splicing. In this article, we summarize our current understanding of the role of phys in the light-mediated pre-mRNA splicing and how that contributes to the regulation of gene expression to promote photomorphogenesis.
Keywords: Arabidopsis; Photomorphogenesis; Phytochrome signaling; Pre-mRNA splicing; Splicing factor.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known copmpeting financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Mathews S, Sharrock RA: Phytochrome gene diversity. Plant Cell Environment 1997, 20:666–671.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

