Gene co-expression network analysis reveals key pathways and hub genes in Chinese cabbage (Brassica rapa L.) during vernalization
- PMID: 33823810
- PMCID: PMC8022416
- DOI: 10.1186/s12864-021-07510-8
Gene co-expression network analysis reveals key pathways and hub genes in Chinese cabbage (Brassica rapa L.) during vernalization
Abstract
Background: Vernalization is a type of low temperature stress used to promote rapid bolting and flowering in plants. Although rapid bolting and flowering promote the reproduction of Chinese cabbages (Brassica rapa L. ssp. pekinensis), this process causes their commercial value to decline. Clarifying the mechanisms of vernalization is essential for its further application. We performed RNA sequencing of gradient-vernalization in order to explore the reasons for the different bolting process of two Chinese cabbage accessions during vernalization.
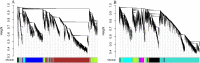
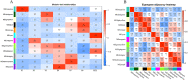
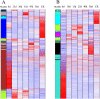
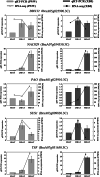
Results: There was considerable variation in gene expression between different-bolting Chinese cabbage accessions during vernalization. Comparative transcriptome analysis and weighted gene co-expression network analysis (WGCNA) were performed for different-bolting Chinese cabbage during different vernalization periods. The biological function analysis and hub gene annotation of highly relevant modules revealed that shoot system morphogenesis and polysaccharide and sugar metabolism caused early-bolting 'XBJ' to bolt and flower faster; chitin, ABA and ethylene-activated signaling pathways were enriched in late-bolting 'JWW'; and leaf senescence and carbohydrate metabolism enrichment were found in the two Chinese cabbage-related modules, indicating that these pathways may be related to bolting and flowering. The high connectivity of hub genes regulated vernalization, including MTHFR2, CPRD49, AAP8, endoglucanase 10, BXLs, GATLs, and WRKYs. Additionally, five genes related to flower development, BBX32 (binds to the FT promoter), SUS1 (increases FT expression), TSF (the closest homologue of FT), PAO and NAC029 (plays a role in leaf senescence), were expressed in the two Chinese cabbage accessions.
Conclusion: The present work provides a comprehensive overview of vernalization-related gene networks in two different-bolting Chinese cabbages during vernalization. In addition, the candidate pathways and hub genes related to vernalization identified here will serve as a reference for breeders in the regulation of Chinese cabbage production.
Keywords: Chinese cabbage; Gradient-vernalization; Hub genes; RNA sequencing; Weighted gene co-expression network analysis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







Similar articles
-
Global Transcriptome and Weighted Gene Co-Expression Network Analyses of Cold Stress Responses in Chinese Cabbage.Genes (Basel). 2025 Jul 20;16(7):845. doi: 10.3390/genes16070845. Genes (Basel). 2025. PMID: 40725500 Free PMC article.
-
ANALYSIS OF GENOMIC DNA METHYLATION AND GENE EXPRESSION IN CHINESE CABBAGE (Brassica rapa L. ssp. pekinensis) AFTER CONTINUOUS SEEDLING BREEDING.Genetika. 2015 Aug;51(8):905-14. doi: 10.7868/s0016675815080111. Genetika. 2015. PMID: 26601490
-
Comparative Transcriptome Analysis of Gene Expression and Regulatory Characteristics Associated with Different Vernalization Periods in Brassica rapa.Genes (Basel). 2020 Apr 5;11(4):392. doi: 10.3390/genes11040392. Genes (Basel). 2020. PMID: 32260536 Free PMC article.
-
[Epigenetics of plant vernalization regulated by non-coding RNAs].Yi Chuan. 2012 Jul;34(7):829-34. doi: 10.3724/sp.j.1005.2012.00829. Yi Chuan. 2012. PMID: 22805208 Review. Chinese.
-
Physiological Control and Genetic Basis of Leaf Curvature and Heading in Brassica rapa L.J Adv Res. 2023 Nov;53:49-59. doi: 10.1016/j.jare.2022.12.010. Epub 2022 Dec 26. J Adv Res. 2023. PMID: 36581197 Free PMC article. Review.
Cited by
-
Single-cell transcriptomic analysis of flowering regulation and vernalization in Chinese cabbage shoot apex.Hortic Res. 2024 Jul 30;11(10):uhae214. doi: 10.1093/hr/uhae214. eCollection 2024 Oct. Hortic Res. 2024. PMID: 39391013 Free PMC article.
-
Weighted gene co-expression network analysis reveals immune evasion related genes in Echinococcus granulosus sensu stricto.Exp Biol Med (Maywood). 2024 Feb 29;249:10126. doi: 10.3389/ebm.2024.10126. eCollection 2024. Exp Biol Med (Maywood). 2024. PMID: 38510493 Free PMC article.
-
Comparison of the transcriptome and metabolome of wheat (Triticum aestivum L.) proteins content during grain formation provides insight.Front Plant Sci. 2024 Jan 18;14:1309678. doi: 10.3389/fpls.2023.1309678. eCollection 2023. Front Plant Sci. 2024. PMID: 38304458 Free PMC article.
-
CRISPR/Cas9-Mediated HY5 Gene Editing Reduces Growth Inhibition in Chinese Cabbage (Brassica rapa) under ER Stress.Int J Mol Sci. 2023 Aug 23;24(17):13105. doi: 10.3390/ijms241713105. Int J Mol Sci. 2023. PMID: 37685921 Free PMC article.
-
Metabolomic and transcriptomic analyses reveal the mechanism of sweet-acidic taste formation during pineapple fruit development.Front Plant Sci. 2022 Sep 8;13:971506. doi: 10.3389/fpls.2022.971506. eCollection 2022. Front Plant Sci. 2022. PMID: 36161024 Free PMC article.
References
-
- Laczi E, Apahidean AS, Apahidean AI, Varga J. Agrobiological Particularities of Chinese Cabbage: Brassica rapa L. ssp. pekinensis (Lour) Hanelt and Brassica rapa L. ssp. chinensis (Lour) Hanelt. Bull Univ Agri Sci Vet Med Cluj-Napoca. 2010;67(1):508.
-
- Eniko L, Alexandru SA, Jolan V, Alexandra IA. Research concerning the bolting of Chinese cabbage (Brassica campestris var. pekinensis (Lour.) Olson) in early crops in polyethylene tunnels. Acta Univ Sapientiae Agri Environ. 2011;3:152–160.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

