Functional genomics of a Spiroplasma associated with the carmine cochineals Dactylopius coccus and Dactylopius opuntiae
- PMID: 33823812
- PMCID: PMC8025503
- DOI: 10.1186/s12864-021-07540-2
Functional genomics of a Spiroplasma associated with the carmine cochineals Dactylopius coccus and Dactylopius opuntiae
Abstract
Background: Spiroplasma is a widely distributed endosymbiont of insects, arthropods, and plants. In insects, Spiroplasma colonizes the gut, hemolymph, and reproductive organs of the host. Previous metagenomic surveys of the domesticated carmine cochineal Dactylopius coccus and the wild cochineal D. opuntiae reported sequences of Spiroplasma associated with these insects. However, there is no analysis of the genomic capabilities and the interaction of this Spiroplasma with Dactylopius.
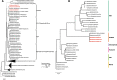
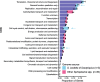
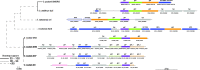
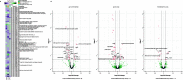
Results: Here we present three Spiroplasma genomes independently recovered from metagenomes of adult males and females of D. coccus, from two different populations, as well as from adult females of D. opuntiae. Single-copy gene analysis showed that these genomes were > 92% complete. Phylogenomic analyses classified these genomes as new members of Spiroplasma ixodetis. Comparative genome analysis indicated that they exhibit fewer genes involved in amino acid and carbon catabolism compared to other spiroplasmas. Moreover, virulence factor-encoding genes (i.e., glpO, spaid and rip2) were found incomplete in these S. ixodetis genomes. We also detected an enrichment of genes encoding the type IV secretion system (T4SS) in S. ixodetis genomes of Dactylopius. A metratranscriptomic analysis of D. coccus showed that some of these T4SS genes (i.e., traG, virB4 and virD4) in addition to the superoxide dismutase sodA of S. ixodetis were overexpressed in the ovaries.
Conclusion: The symbiont S. ixodetis is a new member of the bacterial community of D. coccus and D. opuntiae. The recovery of incomplete virulence factor-encoding genes in S. ixodetis of Dactylopius suggests that this bacterium is a non-pathogenic symbiont. A high number of genes encoding the T4SS, in the S. ixodetis genomes and the overexpression of these genes in the ovary and hemolymph of the host suggest that S. ixodetis use the T4SS to interact with the Dactylopius cells. Moreover, the transcriptional differences of S. ixodetis among the gut, hemolymph and ovary tissues of D. coccus indicate that this bacterium can respond and adapt to the different conditions (e.g., oxidative stress) present within the host. All this evidence proposes that there is a strong interaction and molecular signaling in the symbiosis between S. ixodetis and the carmine cochineal Dactylopius.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Candidatus Dactylopiibacterium carminicum, a Nitrogen-Fixing Symbiont of Dactylopius Cochineal Insects (Hemiptera: Coccoidea: Dactylopiidae).Genome Biol Evol. 2017 Sep 1;9(9):2237-2250. doi: 10.1093/gbe/evx156. Genome Biol Evol. 2017. PMID: 30605507 Free PMC article.
-
Metatranscriptomic Analysis of the Bacterial Symbiont Dactylopiibacterium carminicum from the Carmine Cochineal Dactylopius coccus (Hemiptera: Coccoidea: Dactylopiidae).Life (Basel). 2019 Jan 3;9(1):4. doi: 10.3390/life9010004. Life (Basel). 2019. PMID: 30609847 Free PMC article.
-
Genomic insights into Spiroplasma endosymbionts that induce male-killing and protective phenotypes in the pea aphid.FEMS Microbiol Lett. 2024 Jan 9;371:fnae027. doi: 10.1093/femsle/fnae027. FEMS Microbiol Lett. 2024. PMID: 38632047
-
Spiroplasmas: infectious agents of plants, arthropods and vertebrates.Wien Klin Wochenschr. 1997 Aug 8;109(14-15):604-12. Wien Klin Wochenschr. 1997. PMID: 9286068 Review.
-
The Use of Insect Pigment in Art Works.Insects. 2024 Jul 10;15(7):519. doi: 10.3390/insects15070519. Insects. 2024. PMID: 39057252 Free PMC article. Review.
Cited by
-
Dactylopius opuntiae [Cockerell] Could Be a Source of Antioxidants for the Preservation of Beef Patties.Insects. 2023 Oct 13;14(10):811. doi: 10.3390/insects14100811. Insects. 2023. PMID: 37887823 Free PMC article.
-
The toxins of vertically transmitted Spiroplasma.Front Microbiol. 2023 May 18;14:1148263. doi: 10.3389/fmicb.2023.1148263. eCollection 2023. Front Microbiol. 2023. PMID: 37275155 Free PMC article.
-
Highly transmissible cytoplasmic incompatibility by the extracellular insect symbiont Spiroplasma.iScience. 2022 Apr 29;25(5):104335. doi: 10.1016/j.isci.2022.104335. eCollection 2022 May 20. iScience. 2022. PMID: 35602967 Free PMC article.
-
Intracellular symbiont Symbiodolus is vertically transmitted and widespread across insect orders.ISME J. 2024 Jan 8;18(1):wrae099. doi: 10.1093/ismejo/wrae099. ISME J. 2024. PMID: 38874172 Free PMC article.
-
On the Origins of Symbiotic Fungi in Carmine Cochineals and Their Function in the Digestion of Plant Polysaccharides.Insects. 2024 Oct 9;15(10):783. doi: 10.3390/insects15100783. Insects. 2024. PMID: 39452359 Free PMC article.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

