Single-cell transcriptome atlas and chromatin accessibility landscape reveal differentiation trajectories in the rice root
- PMID: 33824350
- PMCID: PMC8024345
- DOI: 10.1038/s41467-021-22352-4
Single-cell transcriptome atlas and chromatin accessibility landscape reveal differentiation trajectories in the rice root
Abstract
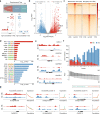
Root development relies on the establishment of meristematic tissues that give rise to distinct cell types that differentiate across defined temporal and spatial gradients. Dissection of the developmental trajectories and the transcriptional networks that underlie them could aid understanding of the function of the root apical meristem in both dicots and monocots. Here, we present a single-cell RNA (scRNA) sequencing and chromatin accessibility survey of rice radicles. By temporal profiling of individual root tip cells we reconstruct continuous developmental trajectories of epidermal cells and ground tissues, and elucidate regulatory networks underlying cell fate determination in these cell lineages. We further identify characteristic processes, transcriptome profiles, and marker genes for these cell types and reveal conserved and divergent root developmental pathways between dicots and monocots. Finally, we demonstrate the potential of the platform for functional genetic studies by using spatiotemporal modeling to identify a rice root meristematic mutant from a cell-specific gene cohort.
Conflict of interest statement
The authors declare no competing interests.
Figures

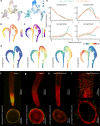


References
-
- Rebouillat J, et al. Molecular genetics of rice root development. Rice. 2009;2:15–34. doi: 10.1007/s12284-008-9016-5. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

