Rare, Protein-Altering Variants in AS3MT and Arsenic Metabolism Efficiency: A Multi-Population Association Study
- PMID: 33826413
- PMCID: PMC8041273
- DOI: 10.1289/EHP8152
Rare, Protein-Altering Variants in AS3MT and Arsenic Metabolism Efficiency: A Multi-Population Association Study
Erratum in
-
Erratum: "Rare, protein-altering variants in AS3MT and arsenic metabolism efficiency: a multi-population association study".Environ Health Perspect. 2021 May;129(5):59003. doi: 10.1289/EHP9488. Epub 2021 May 26. Environ Health Perspect. 2021. PMID: 34038222 Free PMC article. No abstract available.
Abstract
Background: Common genetic variation in the arsenic methyltransferase (AS3MT) gene region is known to be associated with arsenic metabolism efficiency (AME), measured as the percentage of dimethylarsinic acid (DMA%) in the urine. Rare, protein-altering variants in AS3MT could have even larger effects on AME, but their contribution to AME has not been investigated.
Objectives: We estimated the impact of rare, protein-coding variation in AS3MT on AME using a multi-population approach to facilitate the discovery of population-specific and shared causal rare variants.
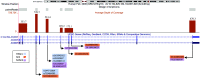
Methods: We generated targeted DNA sequencing data for the coding regions of AS3MT for three arsenic-exposed cohorts with existing data on arsenic species measured in urine: Health Effects of Arsenic Longitudinal Study (HEALS, ), Strong Heart Study (SHS, ), and New Hampshire Skin Cancer Study (NHSCS, ). We assessed the collective effects of rare (allele frequency ), protein-altering AS3MT variants on DMA%, using multiple approaches, including a test of the association between rare allele carrier status (yes/no) and DMA% using linear regression (adjusted for common variants in 10q24.32 region, age, sex, and population structure).
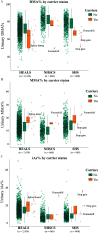
Results: We identified 23 carriers of rare-protein-altering AS3MT variant across all cohorts (13 in HEALS and 5 in both SHS and NHSCS), including 6 carriers of predicted loss-of-function variants. DMA% was 6-10% lower in carriers compared with noncarriers in HEALS [ (95% CI: , )], SHS [ (95% CI: , )], and NHSCS [ (95% CI: , )]. In meta-analyses across cohorts, DMA% was 8.7% lower in carriers [ (95% CI: , )].
Discussion: Rare, protein-altering variants in AS3MT were associated with lower mean DMA%, an indicator of reduced AME. Although a small percentage of the population (0.5-0.7%) carry these variants, they are associated with a 6-10% decrease in DMA% that is consistent across multiple ancestral and environmental backgrounds. https://doi.org/10.1289/EHP8152.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA057494/CA/NCI NIH HHS/United States
- U01 HL041642/HL/NHLBI NIH HHS/United States
- U01 HL041654/HL/NHLBI NIH HHS/United States
- P42 ES010349/ES/NIEHS NIH HHS/United States
- R25 GM109439/GM/NIGMS NIH HHS/United States
- R01 HL109315/HL/NHLBI NIH HHS/United States
- 75N92019D00030/HL/NHLBI NIH HHS/United States
- R01 HL109319/HL/NHLBI NIH HHS/United States
- R21 ES024834/ES/NIEHS NIH HHS/United States
- P30 ES027792/ES/NIEHS NIH HHS/United States
- R01 HL109284/HL/NHLBI NIH HHS/United States
- U01 HL065521/HL/NHLBI NIH HHS/United States
- R01 HL109301/HL/NHLBI NIH HHS/United States
- R01 HL109282/HL/NHLBI NIH HHS/United States
- T32 AG051146/AG/NIA NIH HHS/United States
- R24 ES028532/ES/NIEHS NIH HHS/United States
- R01 ES025216/ES/NIEHS NIH HHS/United States
- P42 ES007373/ES/NIEHS NIH HHS/United States
- 75N92019D00028/HL/NHLBI NIH HHS/United States
- R01 HL090863/HL/NHLBI NIH HHS/United States
- U01 HL041652/HL/NHLBI NIH HHS/United States
- 75N92019D00029/HL/NHLBI NIH HHS/United States
- R01 ES023834/ES/NIEHS NIH HHS/United States
- R24 TW009555/TW/FIC NIH HHS/United States
- R01 CA107431/CA/NCI NIH HHS/United States
- 75N92019D00027/HL/NHLBI NIH HHS/United States
- R35 ES028379/ES/NIEHS NIH HHS/United States
- U01 HL065520/HL/NHLBI NIH HHS/United States
- R01 ES021367/ES/NIEHS NIH HHS/United States
- P20 GM104416/GM/NIGMS NIH HHS/United States
- P30 ES009089/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

