Integration in oncogenes plays only a minor role in determining the in vivo distribution of HIV integration sites before or during suppressive antiretroviral therapy
- PMID: 33826675
- PMCID: PMC8055010
- DOI: 10.1371/journal.ppat.1009141
Integration in oncogenes plays only a minor role in determining the in vivo distribution of HIV integration sites before or during suppressive antiretroviral therapy
Abstract
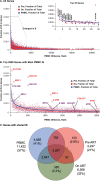
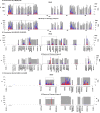
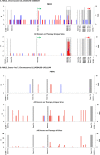
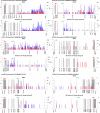
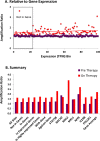
HIV persists during antiretroviral therapy (ART) as integrated proviruses in cells descended from a small fraction of the CD4+ T cells infected prior to the initiation of ART. To better understand what controls HIV persistence and the distribution of integration sites (IS), we compared about 15,000 and 54,000 IS from individuals pre-ART and on ART, respectively, with approximately 395,000 IS from PBMC infected in vitro. The distribution of IS in vivo is quite similar to the distribution in PBMC, but modified by selection against proviruses in expressed genes, by selection for proviruses integrated into one of 7 specific genes, and by clonal expansion. Clones in which a provirus integrated in an oncogene contributed to cell survival comprised only a small fraction of the clones persisting in on ART. Mechanisms that do not involve the provirus, or its location in the host genome, are more important in determining which clones expand and persist.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: JWM is a consultant to Gilead Sciences and Merck Co., Inc., and owns share options in Co-Crystal Pharmaceuticals, Inc. JMC is a member of the Scientific Advisory Board of ROME Therapeutics, Inc. All other authors declare no competing interests.
Figures

References
-
- Joos B, Fischer M, Kuster H, Pillai SK, Wong JK, Boni J, et al.. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(43):16725–30. 10.1073/pnas.0804192105 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

