Light-Activation of DNA-Methyltransferases
- PMID: 33826797
- PMCID: PMC8251764
- DOI: 10.1002/anie.202103945
Light-Activation of DNA-Methyltransferases
Abstract
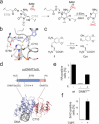
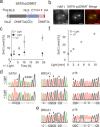
5-Methylcytosine (5mC), the central epigenetic mark of mammalian DNA, plays fundamental roles in chromatin regulation. 5mC is written onto genomes by DNA methyltransferases (DNMT), and perturbation of this process is an early event in carcinogenesis. However, studying 5mC functions is limited by the inability to control individual DNMTs with spatiotemporal resolution in vivo. We report light-control of DNMT catalysis by genetically encoding a photocaged cysteine as a catalytic residue. This enables translation of inactive DNMTs, their rapid activation by light-decaging, and subsequent monitoring of de novo DNA methylation. We provide insights into how cancer-related DNMT mutations alter de novo methylation in vivo, and demonstrate local and tuneable cytosine methylation by light-controlled DNMTs fused to a programmable transcription activator-like effector domain targeting pericentromeric satellite-3 DNA. We further study early events of transcriptome alterations upon DNMT-catalyzed cytosine methylation. Our study sets a basis to dissect the order and kinetics of diverse chromatin-associated events triggered by normal and aberrant DNA methylation.
Keywords: DNA methyltransferases; epigenetics; genetic code expansion; optochemical biology.
© 2021 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Local chromatin microenvironment determines DNMT activity: from DNA methyltransferase to DNA demethylase or DNA dehydroxymethylase.Epigenetics. 2015;10(8):671-6. doi: 10.1080/15592294.2015.1062204. Epigenetics. 2015. PMID: 26098813 Free PMC article. Review.
-
Role of Mammalian DNA Methyltransferases in Development.Annu Rev Biochem. 2020 Jun 20;89:135-158. doi: 10.1146/annurev-biochem-103019-102815. Epub 2019 Dec 9. Annu Rev Biochem. 2020. PMID: 31815535 Review.
-
Rational Manipulation of DNA Methylation by Using Isotopically Reinforced Cytosine.Chembiochem. 2016 Nov 3;17(21):2018-2021. doi: 10.1002/cbic.201600393. Epub 2016 Sep 30. Chembiochem. 2016. PMID: 27595234
-
Genetic Studies on Mammalian DNA Methyltransferases.Adv Exp Med Biol. 2022;1389:111-136. doi: 10.1007/978-3-031-11454-0_5. Adv Exp Med Biol. 2022. PMID: 36350508 Free PMC article.
-
Genetic Studies on Mammalian DNA Methyltransferases.Adv Exp Med Biol. 2016;945:123-150. doi: 10.1007/978-3-319-43624-1_6. Adv Exp Med Biol. 2016. PMID: 27826837 Review.
Cited by
-
Towards site-specific manipulation in cysteine-mediated redox signaling.Chem Sci. 2025 Apr 24;16(21):9049-9055. doi: 10.1039/d5sc02016f. eCollection 2025 May 28. Chem Sci. 2025. PMID: 40321179 Free PMC article. Review.
-
Optogenetics with Atomic Precision─A Comprehensive Review of Optical Control of Protein Function through Genetic Code Expansion.Chem Rev. 2025 Feb 26;125(4):1663-1717. doi: 10.1021/acs.chemrev.4c00224. Epub 2025 Feb 10. Chem Rev. 2025. PMID: 39928721 Free PMC article. Review.
-
Role of DNA methylation transferase in urinary system diseases: From basic to clinical perspectives (Review).Int J Mol Med. 2025 Feb;55(2):19. doi: 10.3892/ijmm.2024.5460. Epub 2024 Nov 22. Int J Mol Med. 2025. PMID: 39575487 Free PMC article. Review.
-
Post-translational Lysine Ac(et)ylation in Bacteria: A Biochemical, Structural, and Synthetic Biological Perspective.Front Microbiol. 2021 Oct 11;12:757179. doi: 10.3389/fmicb.2021.757179. eCollection 2021. Front Microbiol. 2021. PMID: 34721364 Free PMC article. Review.
-
Screening of pivotal oncogenes modulated by DNA methylation in hepatocellular carcinoma and identification of atractylenolide I as an anti-cancer drug.Hum Cell. 2025 May 5;38(4):97. doi: 10.1007/s13577-025-01224-9. Hum Cell. 2025. PMID: 40325252
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

