The chaperone-binding activity of the mitochondrial surface receptor Tom70 protects the cytosol against mitoprotein-induced stress
- PMID: 33826901
- PMCID: PMC7615001
- DOI: 10.1016/j.celrep.2021.108936
The chaperone-binding activity of the mitochondrial surface receptor Tom70 protects the cytosol against mitoprotein-induced stress
Abstract
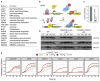
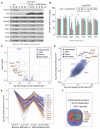
Most mitochondrial proteins are synthesized as precursors in the cytosol and post-translationally transported into mitochondria. The mitochondrial surface protein Tom70 acts at the interface of the cytosol and mitochondria. In vitro import experiments identified Tom70 as targeting receptor, particularly for hydrophobic carriers. Using in vivo methods and high-content screens, we revisit the question of Tom70 function and considerably expand the set of Tom70-dependent mitochondrial proteins. We demonstrate that the crucial activity of Tom70 is its ability to recruit cytosolic chaperones to the outer membrane. Indeed, tethering an unrelated chaperone-binding domain onto the mitochondrial surface complements most of the defects caused by Tom70 deletion. Tom70-mediated chaperone recruitment reduces the proteotoxicity of mitochondrial precursor proteins, particularly of hydrophobic inner membrane proteins. Thus, our work suggests that the predominant function of Tom70 is to tether cytosolic chaperones to the outer mitochondrial membrane, rather than to serve as a mitochondrion-specifying targeting receptor.
Keywords: Tom70; chaperones; mitochondria; outer membrane; protein translocation; proteostasis; prototoxicity.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






References
-
- Altmann K, Dürr M, Westermann B. In: Mitochondria Practical Protocols. Leister D, Herrmann JM, editors. Humana Press; 2007. Saccharomyces cerevisiae as a model organism to study mitochondrial biology. - PubMed
-
- Araiso Y, Tsutsumi A, Qiu J, Imai K, Shiota T, Song J, Lindau C, Wenz LS, Sakaue H, Yunoki K, et al. Structure of the mitochondrial import gate reveals distinct preprotein paths. Nature. 2019;575:395–401. - PubMed
-
- Back R, Dominguez C, Rothé B, Bobo C, Beaufils C, Moréra S, Meyer P, Charpentier B, Branlant C, Allain FH, Manival X. High-resolution structural analysis shows how Tah1 tethers Hsp90 to the R2TP complex. Structure. 2013;21:1834–1847. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

