An in vitro model for assessment of SARS-CoV-2 infectivity by defining the correlation between virus isolation and quantitative PCR value: isolation success of SARS-CoV-2 from oropharyngeal swabs correlates negatively with Cq value
- PMID: 33827618
- PMCID: PMC8025900
- DOI: 10.1186/s12985-021-01542-y
An in vitro model for assessment of SARS-CoV-2 infectivity by defining the correlation between virus isolation and quantitative PCR value: isolation success of SARS-CoV-2 from oropharyngeal swabs correlates negatively with Cq value
Erratum in
-
Correction to: An in vitro model for assessment of SARS‑CoV‑2 infectivity by defining the correlation between virus isolation and quantitative PCR value: isolation success of SARS‑CoV‑2 from oropharyngeal swabs correlates negatively with Cq value.Virol J. 2021 Apr 20;18(1):82. doi: 10.1186/s12985-021-01558-4. Virol J. 2021. PMID: 33879217 Free PMC article. No abstract available.
Abstract
Background: At the beginning of the pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), little was known about its actual rate of infectivity and any COVID-19 patient positive in laboratory testing was supposed to be highly infective and a public health risk factor.
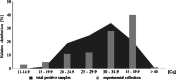
Methods: One hundred oropharyngeal samples were obtained during routine work flow of testing symptomatic persons by quantitative polymerase chain reaction (qPCR) and were inoculated onto cell culture of VeroB4 cells to study the degree of infectivity of SARS-CoV-2 in vitro. Quantification by virus titration and an external standard using synthetic RNA gave the breaking point of infectivity in SARS-CoV-2 in vitro.
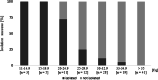
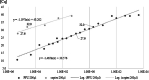
Results: A clear negative correlation (r = - 0.76; p < 0.05) could be asserted between the viral load in quantitative polymerase chain reaction (qPCR) and the probability of a successful isolation in serial isolation experiments of specific oropharyngeal samples positive in qPCR. Quantification by virus titration and an external standard using synthetic RNA indicate a Cq between 27 and 30 in E-gene screening PCR as a breaking point in vitro, where infectivity decreases significantly and isolations become less probable.
Conclusions: This study showed that only the 21% of samples with the highest viral load were infectious enough to transmit the virus in vitro and determined that the dispersion rate in vitro is surprisingly close to those calculated in large retrospective epidemiological studies for SARS-CoV-2. This raises the question of whether this simple in vitro model is suitable to give first insights in dispersion characters of novel or neglected viral pathogens. The statement that SARS-CoV-2 needs at least 40,000 copies to reliably induce infection in vitro is an indication of its transmissibility in Public Health decisions. Applying quantitative PCR systems in diagnosis of SARS-CoV2 can distinguish between patients providing a high risk of transmission and those, where the risk of transmission is probably limited to close and long-lasting contacts.
Keywords: Infectivity; Quantitation of viral infectivity; SARS-CoV-2; Transmission pattern in vitro.
Conflict of interest statement
No consent for publication applicable. This manuscript does not provide any patients’ data nor any animal studies or experiments. This manuscript does not contain any individual person’s data in any form. The authors declare no financial and non-financial conflict of interests. This manuscript has not been submitted to, nor is under review at, another journal or other publishing venue. The authors have no affiliation with any organization with a direct or indirect financial interest in the subject matter discussed in the manuscript.
The authors declare that they have no competing interests.
Figures
Similar articles
-
Assessment of SARS-CoV-2 infectivity of upper respiratory specimens from COVID-19 patients by virus isolation using VeroE6/TMPRSS2 cells.BMJ Open Respir Res. 2021 Feb;8(1):e000830. doi: 10.1136/bmjresp-2020-000830. BMJ Open Respir Res. 2021. PMID: 33627333 Free PMC article.
-
Mechanical transmission of SARS-CoV-2 by house flies.Parasit Vectors. 2021 Apr 20;14(1):214. doi: 10.1186/s13071-021-04703-8. Parasit Vectors. 2021. PMID: 33879234 Free PMC article.
-
A Multiplex One-Step RT-qPCR Protocol to Detect SARS-CoV-2 in NP/OP Swabs and Saliva.Curr Protoc. 2021 May;1(5):e145. doi: 10.1002/cpz1.145. Curr Protoc. 2021. PMID: 34004070 Free PMC article.
-
Viral dynamics in the Upper Respiratory Tract (URT) of SARS-CoV-2.Infez Med. 2020 Dec 1;28(4):486-499. Infez Med. 2020. PMID: 33257622
-
Risk of transmission of severe acute respiratory syndrome coronavirus 2 by transfusion: A literature review.Transfusion. 2020 Dec;60(12):3046-3054. doi: 10.1111/trf.16056. Epub 2020 Sep 1. Transfusion. 2020. PMID: 32798237 Free PMC article. Review.
Cited by
-
SARS-CoV-2 Droplet and Airborne Transmission Heterogeneity.J Clin Med. 2022 May 6;11(9):2607. doi: 10.3390/jcm11092607. J Clin Med. 2022. PMID: 35566733 Free PMC article.
-
SARS-CoV-2 bioaerosol transmission in experimentally infected American mink.Sci Rep. 2025 Jul 1;15(1):22270. doi: 10.1038/s41598-025-08111-1. Sci Rep. 2025. PMID: 40596540 Free PMC article.
-
Evaluation of Four Fully Integrated Molecular Assays for the Detection of Respiratory Viruses during the Co-Circulation of SARS-CoV-2, Influenza and RSV.J Clin Med. 2022 Jul 6;11(14):3942. doi: 10.3390/jcm11143942. J Clin Med. 2022. PMID: 35887705 Free PMC article.
-
Virologic Outcomes with Molnupiravir in Non-hospitalized Adult Patients with COVID-19 from the Randomized, Placebo-Controlled MOVe-OUT Trial.Infect Dis Ther. 2023 Dec;12(12):2725-2743. doi: 10.1007/s40121-023-00891-1. Epub 2023 Nov 23. Infect Dis Ther. 2023. PMID: 37995070 Free PMC article.
-
Generation of quality-controlled SARS-CoV-2 variant stocks.Nat Protoc. 2023 Dec;18(12):3821-3855. doi: 10.1038/s41596-023-00897-6. Epub 2023 Oct 13. Nat Protoc. 2023. PMID: 37833423 Review.
References
-
- Adam D, Wu P, Wong J, Lau E, Tsang T, Cauchemez S, et al. Clustering and superspreading potential of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections in Hong Kong. [PREPRINT (Version 1) available at Research Square. 10.21203/rs3rs-29548/v1. 2020.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

