Prevalence of blaCTX-M Genes in Gram-Negative Bloodstream Isolates across 66 Hospitals in the United States
- PMID: 33827899
- PMCID: PMC8316135
- DOI: 10.1128/JCM.00127-21
Prevalence of blaCTX-M Genes in Gram-Negative Bloodstream Isolates across 66 Hospitals in the United States
Abstract
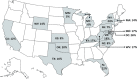
Understanding bacterial species at greatest risk for harboring blaCTX-M genes is necessary to guide antibiotic treatment. We identified the species-specific prevalence of blaCTX-M genes in Gram-negative clinical isolates from the United States. Twenty-four microbiology laboratories representing 66 hospitals using the GenMark Dx ePlex blood culture identification Gram-negative (BCID-GN) panel extracted blood culture results from April 2019 to July 2020. The BCID-GN panel includes 21 Gram-negative targets. Along with identifying blaCTX-M genes, it detects major carbapenemase gene families. A total of 4,209 Gram-negative blood cultures were included. blaCTX-M genes were identified in 462 (11%) specimens. The species-specific prevalence of blaCTX-M genes was as follows: Escherichia coli (16%), Klebsiella pneumoniae (14%), Klebsiella oxytoca (6%), Salmonella spp. (6%), Acinetobacter baumannii (5%), Enterobacter species (3%), Proteus mirabilis (2%), Serratia marcescens (0.6%), and Pseudomonas aeruginosa (0.5%). blaCTX-M prevalence was 26%, 24%, and 22% among participating hospitals in the District of Columbia, New York, and Florida, respectively. Carbapenemase genes were identified in 61 (2%) organisms with the following distribution: blaKPC (59%), blaVIM (16%), blaOXA (10%), blaNDM (8%), and blaIMP (7%). The species-specific prevalence of carbapenemase genes was as follows: A. baumannii (5%), K. pneumoniae (3%), P. mirabilis (3%), Enterobacter species (3%), Citrobacter spp. (3%), P. aeruginosa (2%), E. coli (<1%), K. oxytoca (<1%), and S. marcescens (<1%). Approximately 11% of Gram-negative organisms in our US cohort contain blaCTX-M genes. blaCTX-M genes remain uncommon in organisms beyond E. coli, K. pneumoniae, and K. oxytoca Future molecular diagnostic panels would benefit from the inclusion of plasmid-mediated ampC and SHV and TEM extended-spectrum beta-lactamase (ESBL) targets.
Keywords: AMR; ESBLs; antimicrobial resistance; ceftriaxone.
Copyright © 2021 American Society for Microbiology.
Figures
References
-
- Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2019. www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.... Accessed December 12th, 2020.
-
- The Center for Disease Dynamics, Economics, & Policy. Resistance Map. https://resistancemap.cddep.org/AntibioticResistance.php. Accessed December 12th, 2020.
-
- Hayakawa K, Gattu S, Marchaim D, Bhargava A, Palla M, Alshabani K, Gudur UM, Pulluru H, Bathina P, Sundaragiri PR, Sarkar M, Kakarlapudi H, Ramasamy B, Nanjireddy P, Mohin S, Dasagi M, Datla S, Kuchipudi V, Reddy S, Shahani S, Upputuri V, Marrey S, Gannamani V, Madhanagopal N, Annangi S, Sudha B, Muppavarapu KS, Moshos JA, Lephart PR, Pogue JM, Bush K, Kaye KS. 2013. Epidemiology and risk factors for isolation of Escherichia coli producing CTX-M-type extended-spectrum beta-lactamase in a large U.S. Medical Center. Antimicrob Agents Chemother 57:4010–4018. 10.1128/AAC.02516-12. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

