Cryo-EM structure of the human histamine H1 receptor/Gq complex
- PMID: 33828102
- PMCID: PMC8027608
- DOI: 10.1038/s41467-021-22427-2
Cryo-EM structure of the human histamine H1 receptor/Gq complex
Abstract
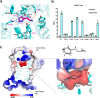
Histamine receptors play important roles in various pathophysiological conditions and are effective targets for anti-allergy treatment, however the mechanism of receptor activation remain elusive. Here, we present the cryo-electron microscopy (cryo-EM) structure of the human H1R in complex with a Gq protein in an active conformation via a NanoBiT tethering strategy. The structure reveals that histamine activates receptor via interacting with the key residues of both transmembrane domain 3 (TM3) and TM6 to squash the binding pocket on the extracellular side and to open the cavity on the intracellular side for Gq engagement in a model of "squash to activate and expand to deactivate". The structure also reveals features for Gq coupling, including the interaction between intracellular loop 2 (ICL2) and the αN-β junction of Gq/11 protein. The detailed analysis of our structure will provide a framework for understanding G-protein coupling selectivity and clues for designing novel antihistamines.
Conflict of interest statement
The authors declare no competing interests.
Figures
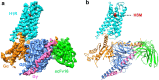

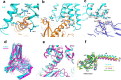
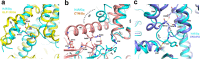
Similar articles
-
Structural basis of ligand recognition and activation of the histamine receptor family.Nat Commun. 2024 Sep 27;15(1):8296. doi: 10.1038/s41467-024-52585-y. Nat Commun. 2024. PMID: 39333117 Free PMC article.
-
Docking and MD study of histamine H4R based on the crystal structure of H1R.J Mol Graph Model. 2013 Feb;39:1-12. doi: 10.1016/j.jmgm.2012.10.003. Epub 2012 Oct 23. J Mol Graph Model. 2013. PMID: 23220277
-
Cryo-EM structures of human bradykinin receptor-Gq proteins complexes.Nat Commun. 2022 Feb 7;13(1):714. doi: 10.1038/s41467-022-28399-1. Nat Commun. 2022. PMID: 35132089 Free PMC article.
-
Structural Analysis of the Histamine H1 Receptor.Handb Exp Pharmacol. 2017;241:21-30. doi: 10.1007/164_2016_10. Handb Exp Pharmacol. 2017. PMID: 27826702 Review.
-
Functional Role of the C-Terminal Amphipathic Helix 8 of Olfactory Receptors and Other G Protein-Coupled Receptors.Int J Mol Sci. 2016 Nov 18;17(11):1930. doi: 10.3390/ijms17111930. Int J Mol Sci. 2016. PMID: 27869740 Free PMC article. Review.
Cited by
-
Coupling and Activation of the β1 Adrenergic Receptor - The Role of the Third Intracellular Loop.J Am Chem Soc. 2024 Oct 3;146(41):28527-37. doi: 10.1021/jacs.4c11250. Online ahead of print. J Am Chem Soc. 2024. PMID: 39359104 Free PMC article.
-
Molecular Mechanisms of Methamphetamine-Induced Addiction via TAAR1 Activation.J Med Chem. 2024 Oct 24;67(20):18593-18605. doi: 10.1021/acs.jmedchem.4c01961. Epub 2024 Oct 2. J Med Chem. 2024. PMID: 39358311 Free PMC article.
-
Structural basis for recognition of antihistamine drug by human histamine receptor.Nat Commun. 2022 Oct 15;13(1):6105. doi: 10.1038/s41467-022-33880-y. Nat Commun. 2022. PMID: 36243875 Free PMC article.
-
Structural insights into the agonists binding and receptor selectivity of human histamine H4 receptor.Nat Commun. 2023 Oct 20;14(1):6538. doi: 10.1038/s41467-023-42260-z. Nat Commun. 2023. PMID: 37863901 Free PMC article.
-
Specific Engineered G Protein Coupling to Histamine Receptors Revealed from Cellular Assay Experiments and Accelerated Molecular Dynamics Simulations.Int J Mol Sci. 2021 Sep 17;22(18):10047. doi: 10.3390/ijms221810047. Int J Mol Sci. 2021. PMID: 34576210 Free PMC article.
References
-
- Fung-Leung WP, Thurmond RL, Ling P, Karlsson L. Histamine H4 receptor antagonists: the new antihistamines? Curr. Opin. Investig. Drugs. 2004;5:1174–1183. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

