VESPER: global and local cryo-EM map alignment using local density vectors
- PMID: 33828103
- PMCID: PMC8027200
- DOI: 10.1038/s41467-021-22401-y
VESPER: global and local cryo-EM map alignment using local density vectors
Abstract
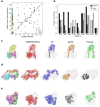
An increasing number of density maps of biological macromolecules have been determined by cryo-electron microscopy (cryo-EM) and stored in the public database, EMDB. To interpret the structural information contained in EM density maps, alignment of maps is an essential step for structure modeling, comparison of maps, and for database search. Here, we developed VESPER, which captures the similarity of underlying molecular structures embedded in density maps by taking local gradient directions into consideration. Compared to existing methods, VESPER achieved substantially more accurate global and local alignment of maps as well as database retrieval.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

