Update of the global distribution of human gammaherpesvirus 8 genotypes
- PMID: 33828146
- PMCID: PMC8026617
- DOI: 10.1038/s41598-021-87038-9
Update of the global distribution of human gammaherpesvirus 8 genotypes
Abstract
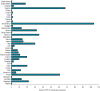
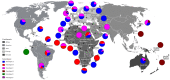
Human gammaherpesvirus 8 (HHV-8) consists of six major clades (A-F) based on the genetic sequence of the open reading frame (ORF)-K1. There are a few conflicting reports regarding the global distribution of the different HHV-8 genotypes. This study aimed to determine the global distribution of the different HHV-8 genotypes based on phylogenetic analysis of the ORF-K1 coding region using sequences published in the GenBank during 1997-2020 and construct a phylogenetic tree using the maximum likelihood algorithm with the GTR + I + G nucleotide substitution model. A total of 550 sequences from 38 countries/origins were analysed in this study. Genotypes A and C had similar global distributions and were prevalent in Africa and Europe. Genotype B was prevalent in Africa. Of the rare genotypes, genotype D was reported in East Asia and Oceania and genotype E in South America, while genotype F was prevalent in Africa. The highest genotypic diversity was reported in the American continent, with Brazil housing five HHV-8 genotypes (A, B, C, E, and F). In this study, we present update of the global distribution of HHV-8 genotypes, providing a basis for future epidemiological and evolutionary studies of HHV-8.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Global Cancer Observatory, WHO. Cancer Today gco.iarc.fr/today/home (2018).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

