Predicting cell-penetrating peptides using machine learning algorithms and navigating in their chemical space
- PMID: 33828175
- PMCID: PMC8027643
- DOI: 10.1038/s41598-021-87134-w
Predicting cell-penetrating peptides using machine learning algorithms and navigating in their chemical space
Abstract
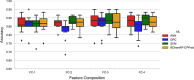
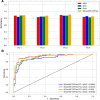
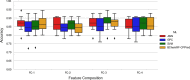
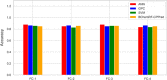
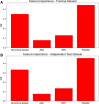
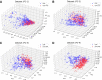
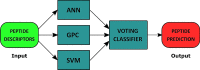
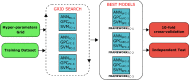
Cell-penetrating peptides (CPPs) are naturally able to cross the lipid bilayer membrane that protects cells. These peptides share common structural and physicochemical properties and show different pharmaceutical applications, among which drug delivery is the most important. Due to their ability to cross the membranes by pulling high-molecular-weight polar molecules, they are termed Trojan horses. In this study, we proposed a machine learning (ML)-based framework named BChemRF-CPPred (beyond chemical rules-based framework for CPP prediction) that uses an artificial neural network, a support vector machine, and a Gaussian process classifier to differentiate CPPs from non-CPPs, using structure- and sequence-based descriptors extracted from PDB and FASTA formats. The performance of our algorithm was evaluated by tenfold cross-validation and compared with those of previously reported prediction tools using an independent dataset. The BChemRF-CPPred satisfactorily identified CPP-like structures using natural and synthetic modified peptide libraries and also obtained better performance than those of previously reported ML-based algorithms, reaching the independent test accuracy of 90.66% (AUC = 0.9365) for PDB, and an accuracy of 86.5% (AUC = 0.9216) for FASTA input. Moreover, our analyses of the CPP chemical space demonstrated that these peptides break some molecular rules related to the prediction of permeability of therapeutic molecules in cell membranes. This is the first comprehensive analysis to predict synthetic and natural CPP structures and to evaluate their chemical space using an ML-based framework. Our algorithm is freely available for academic use at http://comptools.linc.ufpa.br/BChemRF-CPPred .
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Biological Membrane-Penetrating Peptides: Computational Prediction and Applications.Front Cell Infect Microbiol. 2022 Mar 25;12:838259. doi: 10.3389/fcimb.2022.838259. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35402305 Free PMC article. Review.
-
CPPred-RF: A Sequence-based Predictor for Identifying Cell-Penetrating Peptides and Their Uptake Efficiency.J Proteome Res. 2017 May 5;16(5):2044-2053. doi: 10.1021/acs.jproteome.7b00019. Epub 2017 Apr 26. J Proteome Res. 2017. PMID: 28436664
-
Machine-Learning-Based Prediction of Cell-Penetrating Peptides and Their Uptake Efficiency with Improved Accuracy.J Proteome Res. 2018 Aug 3;17(8):2715-2726. doi: 10.1021/acs.jproteome.8b00148. Epub 2018 Jul 2. J Proteome Res. 2018. PMID: 29893128
-
KELM-CPPpred: Kernel Extreme Learning Machine Based Prediction Model for Cell-Penetrating Peptides.J Proteome Res. 2018 Sep 7;17(9):3214-3222. doi: 10.1021/acs.jproteome.8b00322. Epub 2018 Aug 13. J Proteome Res. 2018. PMID: 30032609
-
Empirical comparison and analysis of web-based cell-penetrating peptide prediction tools.Brief Bioinform. 2020 Mar 23;21(2):408-420. doi: 10.1093/bib/bby124. Brief Bioinform. 2020. PMID: 30649170 Review.
Cited by
-
Biological Membrane-Penetrating Peptides: Computational Prediction and Applications.Front Cell Infect Microbiol. 2022 Mar 25;12:838259. doi: 10.3389/fcimb.2022.838259. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35402305 Free PMC article. Review.
-
Research on Plant RNA-Binding Protein Prediction Method Based on Improved Ensemble Learning.Biology (Basel). 2025 Jun 10;14(6):672. doi: 10.3390/biology14060672. Biology (Basel). 2025. PMID: 40563923 Free PMC article.
-
EnDM-CPP: A Multi-view Explainable Framework Based on Deep Learning and Machine Learning for Identifying Cell-Penetrating Peptides with Transformers and Analyzing Sequence Information.Interdiscip Sci. 2025 Sep;17(3):744-769. doi: 10.1007/s12539-024-00673-4. Epub 2024 Dec 23. Interdiscip Sci. 2025. PMID: 39714579
-
TriplEP-CPP: Algorithm for Predicting the Properties of Peptide Sequences.Int J Mol Sci. 2024 Jun 22;25(13):6869. doi: 10.3390/ijms25136869. Int J Mol Sci. 2024. PMID: 38999985 Free PMC article.
-
Screening for effective cell-penetrating peptides with minimal impact on epithelial cells and gut commensals in vitro.Front Pharmacol. 2022 Nov 2;13:1049324. doi: 10.3389/fphar.2022.1049324. eCollection 2022. Front Pharmacol. 2022. PMID: 36408245 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

