Genomic prediction for growth using a low-density SNP panel in dromedary camels
- PMID: 33828208
- PMCID: PMC8027435
- DOI: 10.1038/s41598-021-87296-7
Genomic prediction for growth using a low-density SNP panel in dromedary camels
Abstract
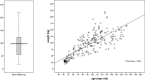
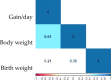
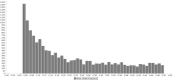
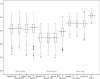
For thousands of years, camels have produced meat, milk, and fiber in harsh desert conditions. For a sustainable development to provide protein resources from desert areas, it is necessary to pay attention to genetic improvement in camel breeding. By using genotyping-by-sequencing (GBS) method we produced over 14,500 genome wide markers to conduct a genome- wide association study (GWAS) for investigating the birth weight, daily gain, and body weight of 96 dromedaries in the Iranian central desert. A total of 99 SNPs were associated with birth weight, daily gain, and body weight (p-value < 0.002). Genomic breeding values (GEBVs) were estimated with the BGLR package using (i) all 14,522 SNPs and (ii) the 99 SNPs by GWAS. Twenty-eight SNPs were associated with birth weight, daily gain, and body weight (p-value < 0.001). Annotation of the genomic region (s) within ± 100 kb of the associated SNPs facilitated prediction of 36 candidate genes. The accuracy of GEBVs was more than 0.65 based on all 14,522 SNPs, but the regression coefficients for birth weight, daily gain, and body weight were 0.39, 0.20, and 0.23, respectively. Because of low sample size, the GEBVs were predicted using the associated SNPs from GWAS. The accuracy of GEBVs based on the 99 associated SNPs was 0.62, 0.82, and 0.57 for birth weight, daily gain, and body weight. This report is the first GWAS using GBS on dromedary camels and identifies markers associated with growth traits that could help to plan breeding program to genetic improvement. Further researches using larger sample size and collaboration of the camel farmers and more profound understanding will permit verification of the associated SNPs identified in this project. The preliminary results of study show that genomic selection could be the appropriate way to genetic improvement of body weight in dromedary camels, which is challenging due to a long generation interval, seasonal reproduction, and lack of records and pedigrees.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Hashim WM, Galal MY, Ali AM, Khalafalla-Abdelmalik AI, Hamid SA, Mohamed KA. Dromedary camels in Sudan, types and sub types, distribution and movement. Int. J. Pharm. Res. Anal. 2015;5:8–12.
-
- Scienski K, et al. Genetic variability in a Holstein population using SNP markers and their use for monitoring mating strategies. Revista Mexicana de Ciencias Pecuarias. 2019;10:643–663. doi: 10.22319/rmcp.v10i3.4842. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

