Alteration of the gut fecal microbiome in children living with HIV on antiretroviral therapy in Yaounde, Cameroon
- PMID: 33828220
- PMCID: PMC8027858
- DOI: 10.1038/s41598-021-87368-8
Alteration of the gut fecal microbiome in children living with HIV on antiretroviral therapy in Yaounde, Cameroon
Abstract
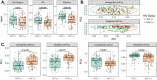
Multiple factors, such as immune disruption, prophylactic co-trimoxazole, and antiretroviral therapy, may influence the structure and function of the gut microbiome of children infected with HIV from birth. In order to understand whether HIV infection altered gut microbiome and to relate changes in microbiome structure and function to immune status, virological response and pediatric ART regimens, we characterized the gut microbiome of 87 HIV-infected and 82 non-exposed HIV-negative children from Yaounde, a cosmopolitan city in Cameroon. We found that children living with HIV had significantly lower alpha diversity in their gut microbiome and altered beta diversity that may not be related to CD4+ T cell count or viral load. There was an increased level of Akkermansia and Faecalibacterium genera and decreased level of Escherichia and other Gamma proteobacteria in children infected with HIV, among other differences. We noted an effect of ethnicity/geography on observed gut microbiome composition and that children on ritonavir-boosted protease inhibitor (PI/r)-based ART had gut microbiome composition that diverged more from HIV-negative controls compared to those on non-nucleoside reverse-transcriptase inhibitors-based ART. Further studies investigating the role of this altered gut microbiome in increased disease susceptibility are warranted for individuals who acquired HIV via mother-to-child transmission.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

