The structure, function and evolution of a complete human chromosome 8
- PMID: 33828295
- PMCID: PMC8099727
- DOI: 10.1038/s41586-021-03420-7
The structure, function and evolution of a complete human chromosome 8
Abstract
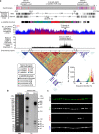
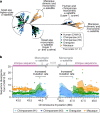
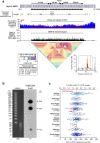
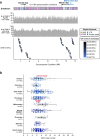
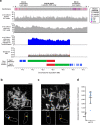
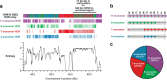
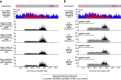
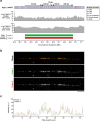
The complete assembly of each human chromosome is essential for understanding human biology and evolution1,2. Here we use complementary long-read sequencing technologies to complete the linear assembly of human chromosome 8. Our assembly resolves the sequence of five previously long-standing gaps, including a 2.08-Mb centromeric α-satellite array, a 644-kb copy number polymorphism in the β-defensin gene cluster that is important for disease risk, and an 863-kb variable number tandem repeat at chromosome 8q21.2 that can function as a neocentromere. We show that the centromeric α-satellite array is generally methylated except for a 73-kb hypomethylated region of diverse higher-order α-satellites enriched with CENP-A nucleosomes, consistent with the location of the kinetochore. In addition, we confirm the overall organization and methylation pattern of the centromere in a diploid human genome. Using a dual long-read sequencing approach, we complete high-quality draft assemblies of the orthologous centromere from chromosome 8 in chimpanzee, orangutan and macaque to reconstruct its evolutionary history. Comparative and phylogenetic analyses show that the higher-order α-satellite structure evolved in the great ape ancestor with a layered symmetry, in which more ancient higher-order repeats locate peripherally to monomeric α-satellites. We estimate that the mutation rate of centromeric satellite DNA is accelerated by more than 2.2-fold compared to the unique portions of the genome, and this acceleration extends into the flanking sequence.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

