Rapid Isolation of Functional ex vivo Human Skin Tissue-Resident Memory T Lymphocytes
- PMID: 33828548
- PMCID: PMC8019735
- DOI: 10.3389/fimmu.2021.624013
Rapid Isolation of Functional ex vivo Human Skin Tissue-Resident Memory T Lymphocytes
Abstract
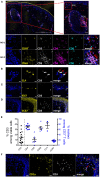
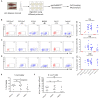
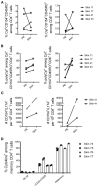
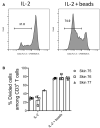
Studies in animal models have shown that skin tissue-resident memory T (TRM) cells provide enhanced and immediate effector function at the site of infection. However, analyses of skin TRM cells in humans have been hindered by the lack of an optimized isolation protocol. Here, we present a combinatorial strategy-the 6-h collagenase IV digestion and gentle tissue dissociation - for rapid and efficient isolation of skin TRM cells with skin tissue-specific immune features. In comparison with paired blood circulating memory T cells, these ex vivo isolated skin T cells express typical TRM cell markers and display higher polyfunctional properties. Moreover, these isolated cells can also be assessed for longer periods of time in ex vivo cultures. Thus, the optimized isolation protocol provides a valuable tool for further understanding of human skin TRM cells, especially for direct comparison with peripheral blood T cells at the same sample collection time.
Keywords: cell isolation; collagenase IV; epitope; gentle tissue dissociation; human skin; tissue-resident memory T cells; yield.
Copyright © 2021 Du, Lenz, Köhler, Zhang, Cendon, Li, Massoud, Wachtlin, Bodo, Hauser, Radbruch and Dong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

