A Rapid and Efficient Screening System for Neutralizing Antibodies and Its Application for SARS-CoV-2
- PMID: 33828563
- PMCID: PMC8019923
- DOI: 10.3389/fimmu.2021.653189
A Rapid and Efficient Screening System for Neutralizing Antibodies and Its Application for SARS-CoV-2
Abstract
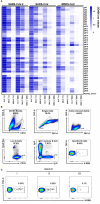
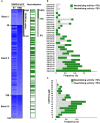
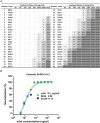
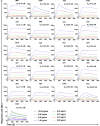
After the pandemic of COVID-19, neutralizing antibodies (NAbs) against SARS-CoV-2 have been developed for the prophylactic and therapeutic purposes. However, few methodologies are described in detail on how to rapidly and efficiently generate effective NAbs to SARS-CoV-2. Here, we integrated and optimized a strategically screening method for NAbs, which has enabled us to obtain SARS-CoV-2 receptor-binding domain (RBD) specific NAbs within 6 days, followed by additional 9 days for antibody production and function analysis. Using this method, we obtained 198 specific Abs against SARS-CoV-2 RBD from the blood samples of COVID-19 convalescent patients, and 96 of them showed neutralizing activity. At least 20% of these NAbs exhibited advanced neutralizing potency and high affinity, with the top two NAbs showing half-maximal inhibitory concentration (IC50) to block authentic SARS-CoV-2 at 9.88 and 11.13 ng/ml, respectively. Altogether, our study provides an effective methodology with high applicable value for discovering potential preventative and therapeutic NAbs for the emerging infectious diseases.
Keywords: SARS-CoV-2; methodology; neutralizing antibodies; receptor-binding domain (RBD); spike protein.
Copyright © 2021 Han, Wang, Li, Hu, Li, Gu, Wang, Shen, Wang, Hu, Wu, Mu, Gong, Chen, Gao, Huang, Long, Luo, Song, Long, Hao, Li, Wu, Xu, Cai, Gao, Zhang, He, Deng, Du, Nai, Wang, Xie, Qu, Huang, Tang and Jin.
Conflict of interest statement
A patent has been filed for some of the antibodies presented here. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Mair-Jenkins J, Saavedra-Campos M, Baillie JK, Cleary P, Khaw FM, Lim WS, et al. The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta-analysis. J Infect Dis (2015) 211:80–90. 10.1093/infdis/jiu396 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

