OsWRKY93 Dually Functions Between Leaf Senescence and in Response to Biotic Stress in Rice
- PMID: 33828575
- PMCID: PMC8019945
- DOI: 10.3389/fpls.2021.643011
OsWRKY93 Dually Functions Between Leaf Senescence and in Response to Biotic Stress in Rice
Abstract
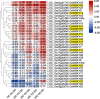
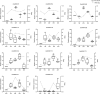
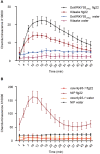
Cross talking between natural senescence and cell death in response to pathogen attack is an interesting topic; however, its action mechanism is kept open. In this study, 33 OsWRKY genes were obtained by screening with leaf aging procedure through RNA-seq dataset, and 11 of them were confirmed a significant altered expression level in the flag leaves during aging by using the reverse transcript quantitative PCR (RT-qPCR). Among them, the OsWRKY2, OsWRKY14, OsWRKY26, OsWRKY69, and OsWRKY93 members exhibited short-term alteration in transcriptional levels in response to Magnaporthe grisea infection. The CRISPR/Cas9-edited mutants of five genes were developed and confirmed, and a significant sensitivity to M. oryzae infection was observed in CRISPR OsWRKY93-edited lines; on the other hand, a significant resistance to M. oryzae infection was shown in the enhanced expression OsWRKY93 plants compared to mock plants; however, enhanced expression of other four genes have no significant affection. Interestingly, ROS accumulation was also increased in OsWRKY93 enhanced plants after flg22 treatment, compared with the controls, suggesting that OsWRKY93 is involved in PAMP-triggered immune response in rice. It indicated that OsWRKY93 was involved in both flag leaf senescence and in response to fungi attack.
Keywords: OsWRKY93; biotic stress; flag leaf; rice; senescence.
Copyright © 2021 Li, Liao, Mei, Pan, Zhang, Zheng, Xie and Miao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Five OsS40 Family Members Are Identified as Senescence-Related Genes in Rice by Reverse Genetics Approach.Front Plant Sci. 2021 Sep 3;12:701529. doi: 10.3389/fpls.2021.701529. eCollection 2021. Front Plant Sci. 2021. PMID: 34539694 Free PMC article.
-
Functional inactivation of OsGCNT induces enhanced disease resistance to Xanthomonas oryzae pv. oryzae in rice.BMC Plant Biol. 2018 Nov 1;18(1):264. doi: 10.1186/s12870-018-1489-9. BMC Plant Biol. 2018. PMID: 30382816 Free PMC article.
-
A proteomic insight into the MSP1 and flg22 induced signaling in Oryza sativa leaves.J Proteomics. 2019 Mar 30;196:120-130. doi: 10.1016/j.jprot.2018.04.015. Epub 2018 Jun 30. J Proteomics. 2019. PMID: 29970347
-
Characterization of S40-like proteins and their roles in response to environmental cues and leaf senescence in rice.BMC Plant Biol. 2019 May 2;19(1):174. doi: 10.1186/s12870-019-1767-1. BMC Plant Biol. 2019. PMID: 31046677 Free PMC article.
-
Osa-miR7695 enhances transcriptional priming in defense responses against the rice blast fungus.BMC Plant Biol. 2019 Dec 18;19(1):563. doi: 10.1186/s12870-019-2156-5. BMC Plant Biol. 2019. PMID: 31852430 Free PMC article.
Cited by
-
DkWRKY transcription factors enhance persimmon resistance to Colletotrichum horii by promoting lignin accumulation through DkCAD1 promotor interaction.Stress Biol. 2024 Feb 26;4(1):17. doi: 10.1007/s44154-024-00154-0. Stress Biol. 2024. PMID: 38407659 Free PMC article.
-
The atypical Dof transcriptional factor OsDes1 contributes to stay-green, grain yield, and disease resistance in rice.Sci Adv. 2024 Aug 23;10(34):eadp0345. doi: 10.1126/sciadv.adp0345. Epub 2024 Aug 23. Sci Adv. 2024. PMID: 39178266 Free PMC article.
-
Use of CRISPR Technology in Gene Editing for Tolerance to Biotic Factors in Plants: A Systematic Review.Curr Issues Mol Biol. 2024 Oct 2;46(10):11086-11123. doi: 10.3390/cimb46100659. Curr Issues Mol Biol. 2024. PMID: 39451539 Free PMC article. Review.
-
Current Understanding of Leaf Senescence in Rice.Int J Mol Sci. 2021 Apr 26;22(9):4515. doi: 10.3390/ijms22094515. Int J Mol Sci. 2021. PMID: 33925978 Free PMC article. Review.
-
Genome and Transcriptome-Wide Analysis of OsWRKY and OsNAC Gene Families in Oryza sativa and Their Response to White-Backed Planthopper Infestation.Int J Mol Sci. 2022 Dec 6;23(23):15396. doi: 10.3390/ijms232315396. Int J Mol Sci. 2022. PMID: 36499722 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

