CEM500K, a large-scale heterogeneous unlabeled cellular electron microscopy image dataset for deep learning
- PMID: 33830015
- PMCID: PMC8032397
- DOI: 10.7554/eLife.65894
CEM500K, a large-scale heterogeneous unlabeled cellular electron microscopy image dataset for deep learning
Abstract
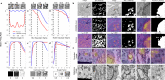
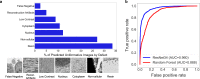
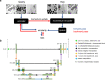
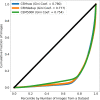
Automated segmentation of cellular electron microscopy (EM) datasets remains a challenge. Supervised deep learning (DL) methods that rely on region-of-interest (ROI) annotations yield models that fail to generalize to unrelated datasets. Newer unsupervised DL algorithms require relevant pre-training images, however, pre-training on currently available EM datasets is computationally expensive and shows little value for unseen biological contexts, as these datasets are large and homogeneous. To address this issue, we present CEM500K, a nimble 25 GB dataset of 0.5 × 106 unique 2D cellular EM images curated from nearly 600 three-dimensional (3D) and 10,000 two-dimensional (2D) images from >100 unrelated imaging projects. We show that models pre-trained on CEM500K learn features that are biologically relevant and resilient to meaningful image augmentations. Critically, we evaluate transfer learning from these pre-trained models on six publicly available and one newly derived benchmark segmentation task and report state-of-the-art results on each. We release the CEM500K dataset, pre-trained models and curation pipeline for model building and further expansion by the EM community. Data and code are available at https://www.ebi.ac.uk/pdbe/emdb/empiar/entry/10592/ and https://git.io/JLLTz.
Keywords: cell biology; computational biology; deep learning; electron microscopy; image dataset; neural network; none; segmentation; systems biology; vEM.
Conflict of interest statement
RC, KN No competing interests declared
Figures






Similar articles
-
Instance segmentation of mitochondria in electron microscopy images with a generalist deep learning model trained on a diverse dataset.Cell Syst. 2023 Jan 18;14(1):58-71.e5. doi: 10.1016/j.cels.2022.12.006. Cell Syst. 2023. PMID: 36657391 Free PMC article.
-
RETINA: Reconstruction-based pre-trained enhanced TransUNet for electron microscopy segmentation on the CEM500K dataset.PLoS Comput Biol. 2025 May 28;21(5):e1013115. doi: 10.1371/journal.pcbi.1013115. eCollection 2025 May. PLoS Comput Biol. 2025. PMID: 40435368 Free PMC article.
-
Segmentation in large-scale cellular electron microscopy with deep learning: A literature survey.Med Image Anal. 2023 Oct;89:102920. doi: 10.1016/j.media.2023.102920. Epub 2023 Aug 6. Med Image Anal. 2023. PMID: 37572414 Review.
-
Deep learning based domain adaptation for mitochondria segmentation on EM volumes.Comput Methods Programs Biomed. 2022 Jul;222:106949. doi: 10.1016/j.cmpb.2022.106949. Epub 2022 Jun 14. Comput Methods Programs Biomed. 2022. PMID: 35753105
-
Benchmarking of deep learning algorithms for 3D instance segmentation of confocal image datasets.PLoS Comput Biol. 2022 Apr 14;18(4):e1009879. doi: 10.1371/journal.pcbi.1009879. eCollection 2022 Apr. PLoS Comput Biol. 2022. PMID: 35421081 Free PMC article. Review.
Cited by
-
Modular segmentation, spatial analysis and visualization of volume electron microscopy datasets.Nat Protoc. 2024 May;19(5):1436-1466. doi: 10.1038/s41596-024-00957-5. Epub 2024 Feb 29. Nat Protoc. 2024. PMID: 38424188 Review.
-
Instance segmentation of mitochondria in electron microscopy images with a generalist deep learning model trained on a diverse dataset.Cell Syst. 2023 Jan 18;14(1):58-71.e5. doi: 10.1016/j.cels.2022.12.006. Cell Syst. 2023. PMID: 36657391 Free PMC article.
-
Deep learning-driven automated mitochondrial segmentation for analysis of complex transmission electron microscopy images.Sci Rep. 2025 May 30;15(1):19076. doi: 10.1038/s41598-025-03311-1. Sci Rep. 2025. PMID: 40447684 Free PMC article.
-
How innovations in methodology offer new prospects for volume electron microscopy.J Microsc. 2022 Sep;287(3):114-137. doi: 10.1111/jmi.13134. Epub 2022 Jul 27. J Microsc. 2022. PMID: 35810393 Free PMC article. Review.
-
Morphomics via next-generation electron microscopy.J Mol Cell Biol. 2024 Apr 10;15(12):mjad081. doi: 10.1093/jmcb/mjad081. J Mol Cell Biol. 2024. PMID: 38148118 Free PMC article. Review.
References
-
- Buhmann J. Automatic detection of synaptic partners in a Whole-Brain Drosophila EM Dataset. bioRxiv. 2019 doi: 10.1101/2019.12.12.874172. - DOI
-
- Carion N, Massa F, Synnaeve G, Usunier N, Kirillov A, Zagoruyko S. End-to-End object detection with transformers. arXiv. 2020 https://arxiv.org/abs/2005.12872
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

