Novel clostridial cell-surface hemicellulose-binding CBM3 proteins
- PMID: 33830074
- PMCID: PMC8034430
- DOI: 10.1107/S2053230X21002764
Novel clostridial cell-surface hemicellulose-binding CBM3 proteins
Abstract
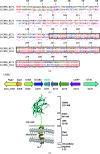
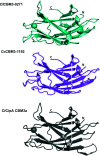
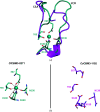
A novel member of the family 3 carbohydrate-binding modules (CBM3s) is encoded by a gene (Cthe_0271) in Clostridium thermocellum which is the most highly expressed gene in the bacterium during its growth on several types of biomass substrates. Surprisingly, CtCBM3-0271 binds to at least two different types of xylan, instead of the common binding of CBM3s to cellulosic substrates. CtCBM3-0271 was crystallized and its three-dimensional structure was solved and refined to a resolution of 1.8 Å. In order to learn more about the role of this type of CBM3, a comparative study with its orthologue from Clostridium clariflavum (encoded by the Clocl_1192 gene) was performed, and the three-dimensional structure of CcCBM3-1192 was determined to 1.6 Å resolution. Carbohydrate binding by CcCBM3-1192 was found to be similar to that by CtCBM3-0271; both exhibited binding to xylan rather than to cellulose. Comparative structural analysis of the two CBM3s provided a clear functional correlation of structure and binding, in which the two CBM3s lack the required number of binding residues in their cellulose-binding strips and thus lack cellulose-binding capabilities. This is an enigma, as CtCBM3-0271 was reported to be a highly expressed protein when the bacterium was grown on cellulose. An additional unexpected finding was that CcCBM3-1192 does not contain the calcium ion that was considered to play a structural stabilizing role in the CBM3 family. Despite the lack of calcium, the five residues that form the calcium-binding site are conserved. The absence of calcium results in conformational changes in two loops of the CcCBM3-1192 structure. In this context, superposition of the non-calcium-binding CcCBM3-1192 with CtCBM3-0271 and other calcium-binding CBM3s reveals a much broader two-loop region in the former compared with CtCBM3-0271.
Keywords: CBM; Clostridium clariflavum; Clostridium thermocellum; calcium binding; cellulosome; crystal structure.
Figures


References
-
- Bayer, E. A., Shimon, L. J. W., Shoham, Y. & Lamed, R. (1998). J. Struct. Biol. 124, 221–234. - PubMed
-
- Bellapadrona, G., Chiaraluce, R., Consalvi, V., Ilari, A., Stefanini, S. & Chiancone, E. (2007). Proteins, 66, 975–983. - PubMed
-
- Benhar, I. (2001). Biotechnol. Adv. 19, 1–33. - PubMed
-
- Burstein, T., Shulman, M., Jindou, S., Petkun, S., Frolow, F., Shoham, Y., Bayer, E. A. & Lamed, R. (2009). FEBS Lett. 583, 879–884. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

